Using Different Regression Methods to Estimate Leaf Nitrogen Content in Rice by Fusing Hyperspectral LiDAR Data and Laser-Induced Chlorophyll Fluorescence Data
Abstract
:1. Introduction
2. Materials and Experiments
2.1. Reflectance Spectrum of Rice Leaf
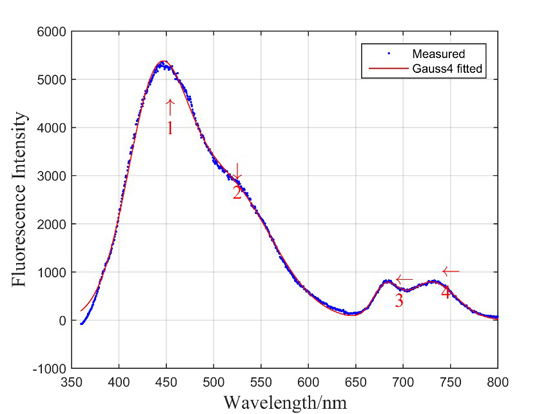
2.2. Laser-Induced Fluorescence of Rice Leaf (LIF)
2.3. Materials and the Design of Experiments
3. Data Analysis
3.1. Regression Methods
3.1.1. Support Vector Machines (SVMs) Regression
3.1.2. Partial Least Squares (PLS)
3.1.3. Artificial Neural Networks (ANNs)
3.2. Re-Ranking Wavelength Method
4. Results and Discussion
4.1. LNC Estimation Using Reflectance Spectra Based on Different Regression Methods
4.2. LNC Estimation Using Reflectance Spectrum and Reflectance + Fluorescence Spectrum
4.3. Band Combinations Based on HSL Data for Rice LNC Estimation
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LNC | Leaf Nitrogen Content |
| LIF | Laser-induced Fluorescence |
| HSL | Hyperspectral LiDAR |
| SVMs | Support Vector Machines |
| PLSR | Partial Least Squares Regression |
| PLS | Partial Least Squares |
| ANNs | Artificial Neural Networks |
| RBF-NNs | Radial Basis Function Neural Networks |
| BP-NNs | Back Propagation Neural Networks |
References
- Johnson, L.F. Nitrogen influence on fresh-leaf NIR spectra. Remote Sens. Environ. 2001, 78, 314–320. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Peterson, D.L. Estimating the foliar biochemical concentration of leaves with reflectance spectrometry: Testing the Kokaly and Clark methodologies. Remote Sens. Environ. 2001, 76, 349–359. [Google Scholar] [CrossRef]
- Kergoat, L.; Lafont, S.; Arneth, A.; Le Dantec, V.; Saugier, B. Nitrogen controls plant canopy light-use efficiency in temperate and boreal ecosystems. J. Geophys. Res. Biogeosci. 2008, 113, 1–19. [Google Scholar] [CrossRef]
- Yao, X.; Huang, Y.; Shang, G.Y.; Zhou, C.; Cheng, T.; Tian, Y.C.; Cao, W.X.; Zhu, Y. Evaluation of Six Algorithms to Monitor Wheat Leaf Nitrogen Concentration. Remote Sens. 2015, 7, 14939–14966. [Google Scholar] [CrossRef]
- Nevalainen, O.; Hakala, T.; Suomalainen, J.; Mäkipää, R.; Peltoniemi, M.; Krooks, A.; Kaasalainen, S. Fast and nondestructive method for leaf level chlorophyll estimation using hyperspectral LiDAR. Agric. Forest Meteorol. 2014, 198, 250–258. [Google Scholar] [CrossRef]
- Woodhouse, I.H.; Nichol, C.; Sinclair, P.; Jack, J.; Morsdorf, F.; Malthus, T.J. A multispectral canopy LiDAR demonstrator project. IEEE Geosci. Remote Sens. Lett. 2011, 8, 839–843. [Google Scholar] [CrossRef]
- Niu, Z.; Xu, Z.; Sun, G.; Huang, W.; Wang, L.; Feng, M.; Li, W.; He, W.B.; Gao, S. Design of a New Multispectral Waveform LiDAR Instrument to Monitor Vegetation. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1506–1510. [Google Scholar]
- Wallace, A.; Nichol, C.; Woodhouse, I. Recovery of forest canopy parameters by inversion of multispectral LiDAR data. Remote Sens. 2012, 4, 509–531. [Google Scholar] [CrossRef]
- Apostol, S.; Viau, A.A.; Tremblay, N. A comparison of multiwavelength laser-induced fluorescence parameters for the remote sensing of nitrogen stress in field-cultivated corn. Can. J. Remote Sens. 2007, 33, 150–161. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Zarco-Tejada, P.J.; Miller, J.R.; Mohammed, G.H.; Noland, T.L. Chlorophyll fluorescence effects on vegetation apparent reflectance: I. Leaf-level measurements and model simulation. Remote Sens. Environ. 2000, 74, 582–595. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Wood, F.M.; McMurtrey, J.E.; Newcomb, W.W. Laser-induced fluorescence of green plants. 1: A technique for the remote detection of plant stress and species differentiation. Appl. Opt. 1984, 23, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Subhash, N.; Mohanan, C. Laser-induced red chlorophyll fluorescence signatures as nutrient stress indicator in rice plants. Remote Sens. Environ. 1994, 47, 45–50. [Google Scholar] [CrossRef]
- Alekseyev, V.; Babichenko, S.; Sobolev, I. Control and signal processing system of hyperspectral FLS LiDAR. In Proceedings of the 2008 11th International Biennial Baltic Electronics Conference, Tallinn, Estonia, 6–8 October 2008; pp. 349–352.
- Haboudane, D.; Tremblay, N.; Miller, J.R.; Vigneault, P. Remote estimation of crop chlorophyll content using spectral indices derived from hyperspectral data. Geosci. Remote Sens. IEEE Trans. 2008, 46, 423–437. [Google Scholar] [CrossRef]
- Schlemmer, A.G.M.; Schepers, J.; Ferguson, R.; Peng, Y.; Shanahan, J.; Rundquist, D. Remote estimation of nitrogen and chlorophyll contents in maize at leaf and canopy levels. Int. J. Appl. Earth Obs. Geoinformation 2013, 25, 47–54. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Berjón, A.; López-Lozano, R.; Miller, J.; Martín, P.; Cachorro, V.; González, M.R.; Frutos, D.A. Assessing vineyard condition with hyperspectral indices: Leaf and canopy reflectance simulation in a row-structured discontinuous canopy. Remote Sens. Environ. 2005, 99, 271–287. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Miehe, J.A. Fluorescence imaging as a diagnostic tool for plant stress. Trends Plant Sci. 1997, 2, 316–320. [Google Scholar] [CrossRef]
- Stober, F.; Lichtenthaler, H.K. Characterization of the laser-induced blue, green and red fluorescence signatures of leaves of wheat and soybean grown under different irradiance. Physiol. Plant. 1993, 88, 696–704. [Google Scholar] [CrossRef]
- Hansen, P.; Schjoerring, J. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Song, S.; Gong, W.; Zhu, B.; Huang, X. Wavelength selection and spectral discrimination for paddy rice, with laboratory measurements of hyperspectral leaf reflectance. ISPRS J. Photogramm. Remote Sens. 2011, 66, 672–682. [Google Scholar] [CrossRef]
- Du, L.; Shi, S.; Yang, J.; Sun, J.; Gong, W. Estimation of leaf nitrogen content of rice using spectral indices and partial least squares regression based on hyperspectral LIDAR data. Unpublished Work. 2016. [Google Scholar]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Yu, K.; Li, F.; Martin, L.G.; Miao, Y.; Georg, B.; Chen, X.P. Remotely detecting canopy nitrogen concentration and uptake of paddy rice in the Northeast China Plain. ISPRS J. Photogramm. Remote Sens. 2013, 78, 102–115. [Google Scholar] [CrossRef]
- Farifteh, J.; Van der Meer, F.; Atzberger, C.; Carranza, E.J.M. Quantitative analysis of salt-affected soil reflectance spectra: A comparison of two adaptive methods (PLSR and ANN). Remote Sens. Environ. 2007, 110, 59–78. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Luis, G.; Jordi, M.; Joan, V.; Julia, A.; Javier, C. Retrieval of oceanic chlorophyll concentration with relevance vector machines. Remote Sens. Environ. 2006, 105, 23–33. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. An assessment of the effectiveness of decision tree methods for land cover classification. Remote Sens. Environ. 2003, 86, 554–565. [Google Scholar] [CrossRef]
- Mantero, P.; Moser, G.; Serpico, S.B. Partially supervised classification of remote sensing images through SVM-based probability density estimation. Geosci. Remote Sens. IEEE Trans. 2005, 43, 559–570. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Samanta, B.; Al-Balushi, K.; Al-Araimi, S. Artificial neural networks and support vector machines with genetic algorithm for bearing fault detection. Eng. Appl. Artif. Intell. 2003, 16, 657–665. [Google Scholar] [CrossRef]
- Römer, C.; Bürling, K.; Hunsche, M.; Rumpf, T.; Noga, G.; Plümer, L. Robust fitting of fluorescence spectra for pre-symptomatic wheat leaf rust detection with support vector machines. Comput. Electron. Agric. 2011, 79, 180–188. [Google Scholar] [CrossRef]
- Gill, M.K.; Asefa, T.; Mariush, W.K.; Mckee, M. Soil moisture prediction using support vector machines 1. J. Am. Water Resour. Assoc. 2006, 42, 1033–1046. [Google Scholar] [CrossRef]
- Yang, X.; Huang, J.; Wu, Y.P.; Wang, W.J.; Wang, P.; Wang, X.; Huete, A.R. Estimating biophysical parameters of rice with remote sensing data using support vector machines. Sci. China Life Sci. 2011, 54, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Gong, W.; Shi, S.; Yang, J.; Sun, J.; Zhu, B.; Song, S. Estimation of rice leaf nitrogen contents based on hyperspectral LIDAR. Int. J. Appl. Earth Obs. Geoinform. 2016, 44, 136–143. [Google Scholar] [CrossRef]
- Dahn, H.; Gunther, K.; Ludeker, W. Characterization of drought stress of maize and wheat canopies by means of spectral resolved laser induced fluorescence. EARSeL Adv. Remote Sens. 1992, 1, 12–19. [Google Scholar]
- Günther, K.; Dahn, H.G.; Lüdeker, W. Remote sensing vegetation status by laser-induced fluorescence. Remote Sens. Environ. 1994, 47, 10–17. [Google Scholar] [CrossRef]
- Yang, J.; Gong, W.; Shi, S.; Du, L.; Sun, J.; Zhu, B.; Ma, Y.; Song, S. Vegetation identification based on characteristics of fluorescence spectral spatial distribution. RSC Adv. 2015, 5, 56932–56935. [Google Scholar] [CrossRef]
- Stocker, T.; Qin, D.; Plattner, G.; Tignor, M.; Allen, S.; Boschung, J. Climate Change 2013: The Physical Science Basis; Intergovernmental Panel on Climate Change: Geneva, Switzerland, 2013. [Google Scholar]
- Wutzke, K.D.; Heine, W. A century of Kjeldahl’s nitrogen determination. Z. Med. Lab. 1984, 26, 383–388. [Google Scholar]
- Vapnick, V.N. Statistical Learning Theory; John Wiley and Sons Inc.: New York, NY, USA, 1998. [Google Scholar]
- Wold, S.; Ruhe, A.; Wold, H.; Dunn, W.J., III. The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J. Sci. Statist. Comput. 1984, 5, 735–743. [Google Scholar] [CrossRef]
- Paya, B.; Esat, I.; Badi, M. Artificial neural network based fault diagnostics of rotating machinery using wavelet transforms as a preprocessor. Mech. Syst. Process. 1997, 11, 751–765. [Google Scholar] [CrossRef]
- Huang, R.; He, M. Band selection based on feature weighting for classification of hyperspectral data. IEEE Geosci. Remote Sens. Lett. 2005, 2, 156–159. [Google Scholar] [CrossRef]
- Eitel, J.U.; Magney, T.S.; Vierling, L.A.; Brown, T.T.; Huggins, D.R. LiDAR based biomass and crop nitrogen estimates for rapid, non-destructive assessment of wheat nitrogen status. Field Crops Res. 2014, 159, 21–32. [Google Scholar] [CrossRef]
- Erdle, K.; Mistele, B.; Schmidhalter, U. Comparison of active and passive spectral sensors in discriminating biomass parameters and nitrogen status in wheat cultivars. Field Crops Res. 2011, 124, 74–84. [Google Scholar] [CrossRef]
- Živčák, M.; Olšovská, K.; Slamka, P.; Galambošová, J.; Rataj, V.; Shao, H.B.; Brestič, M. Application of chlorophyll fluorescence performance indices to assess the wheat photosynthetic functions influenced by nitrogen deficiency. Plant Soil Environ. 2014, 60, 210–215. [Google Scholar]
- Yang, J.; Shi, S.; Gong, W.; Du, L.; Ma, Y.; Zhu, B.; Song, S. Application of fluorescence spectrum to precisely inverse paddy rice nitrogen content. Plant Soil Environ. 2015, 61, 182–188. [Google Scholar] [CrossRef]
- Tian, Y.C.; Gu, K.J.; Chu, X.; Yao, X.; Cao, W.X.; Zhu, Y. Comparison of different hyperspectral vegetation indices for canopy leaf nitrogen concentration estimation in rice. Plant Soil 2014, 376, 193–209. [Google Scholar] [CrossRef]
- Novoa, R.; Loomis, R. Nitrogen and plant production. Plant Soil 1981, 58, 177–204. [Google Scholar] [CrossRef]
- Hamid, A. Efficiency of N uptake by wheat, as affected by time and rate of application, using N15-labelled ammonium sulphate and sodium nitrate. Plant Soil 1972, 37, 389–394. [Google Scholar] [CrossRef]



| Models | Stages | Reflectance | Reflectance + Fluorescence (Peaks or Index) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Rmax2 | RMSE | Parameters | Wavelengths (nm) | Rmax2 | RMSE | Parameters | Wavelengths (nm) | ||
| SVMs | booting | 0.963 | 0.093 | c 1 = 0.707 | 754 694 742 814 790 634 874 886 610 718 706 778 622 586 550 | Peaks 3 0.966 | 0.092 | c = 0.707 γ = 0.008 | 450 685 |
| γ 2 = 1024 | Index 4 - | - | - | - | |||||
| heading | 0.93 | 0.104 | c = 0.707 | 910 634 562 886 706 550 874 826 646 | Peaks 0.937 | 0.099 | c = 0.5 γ = 0.000977 | 685 740 | |
| γ = 1024 | Index 0.899 | 0.103 | c = 0.707 γ = 1024 | 450 & 740 | |||||
| BP-NNs | booting | 0.889 | 0.115 | l 5 = 10 | 682 766 754 742 826 862 634 898 598 610 718 | Peaks 0.882 | 0.129 | l = 10 | 510 |
| Index 0.893 | 0.09 | 450 & 740 | |||||||
| heading | 0.856 | 0.099 | l = 10 | 910 802 670 538 706 550 874 898 586 862 658 574 | Peaks 0.853 | 0.117 | l = 10 | 450 740 | |
| Index - | - | - | |||||||
| RBF-NNs | booting | 0.979 | 0.052 | s 6 = 5 | 682 766 850 754 742 826 814 790 898 | Peaks 0.959 | 0.076 | s = 5 | 685 |
| Index 0.948 | 0.079 | 450 & 685 | |||||||
| heading | 0.957 | 0.063 | s = 5 | 910 634 598 886 538 790 622 898 646 586 | Peaks 0.918 | 0.068 | s = 5 | 450 510 | |
| Index 0.927 | 0.071 | 450 & 685 | |||||||
| PLS | booting | 0.845 | 0.296 | PCs 7 = 6 | 682 850 754 838 802 634 658 730 646 | Peaks 0.832 | 0.279 | PCs = 4 | 450 685 |
| Index 0.887 | 0.26 | PCs = 5 | 450 & 740 | ||||||
| heading | 0.811 | 0.203 | PCs = 6 | 910 589 802 838 706 622 874 646 586 682 658 610 694 | Peaks 0.867 | 0.196 | PCs = 5 | 450 685 | |
| Index 0.874 | 0.194 | PCs = 5 | 450 & 740 | ||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, L.; Shi, S.; Yang, J.; Sun, J.; Gong, W. Using Different Regression Methods to Estimate Leaf Nitrogen Content in Rice by Fusing Hyperspectral LiDAR Data and Laser-Induced Chlorophyll Fluorescence Data. Remote Sens. 2016, 8, 526. https://doi.org/10.3390/rs8060526
Du L, Shi S, Yang J, Sun J, Gong W. Using Different Regression Methods to Estimate Leaf Nitrogen Content in Rice by Fusing Hyperspectral LiDAR Data and Laser-Induced Chlorophyll Fluorescence Data. Remote Sensing. 2016; 8(6):526. https://doi.org/10.3390/rs8060526
Chicago/Turabian StyleDu, Lin, Shuo Shi, Jian Yang, Jia Sun, and Wei Gong. 2016. "Using Different Regression Methods to Estimate Leaf Nitrogen Content in Rice by Fusing Hyperspectral LiDAR Data and Laser-Induced Chlorophyll Fluorescence Data" Remote Sensing 8, no. 6: 526. https://doi.org/10.3390/rs8060526
APA StyleDu, L., Shi, S., Yang, J., Sun, J., & Gong, W. (2016). Using Different Regression Methods to Estimate Leaf Nitrogen Content in Rice by Fusing Hyperspectral LiDAR Data and Laser-Induced Chlorophyll Fluorescence Data. Remote Sensing, 8(6), 526. https://doi.org/10.3390/rs8060526