3D Imaging of Greenhouse Plants with an Inexpensive Binocular Stereo Vision System
Abstract
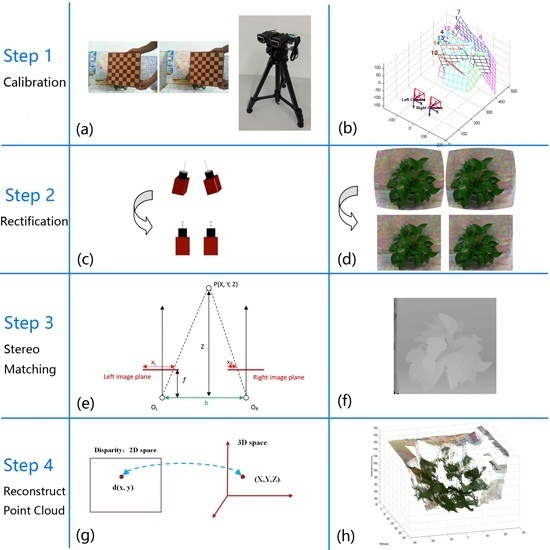
:1. Introduction
- (i)
- To build a highly feasible stereo platform that does not place harsh limitations on hardware and imaged objects. We established an inexpensive (less than 70 USD) and portable binocular stereo vision platform that can be controlled by a laptop. The platform is suitable for 3D imaging of many kinds of plants in different environments such as an indoor lab, open field, and greenhouse.
- (ii)
- To design an effective stereo matching algorithm that not only works on a binocular stereo system, but is also potentially applicable to a SFM-MVS system or any multi-view imaging systems. Improvements in the ASW stereo matching framework are made by replacing the TAD measure with AD-Census measure. The proposed algorithm shows superior results in comparison with the original ASW and several popular energy-based stereo matching algorithms.
- (iii)
- To perform error analysis of the stereo platform from both theoretical and experimental sides. Detailed investigation of the accuracy of the proposed platform under different parametric configurations (e.g., baseline) is provided. For an object that is about 800 mm away from our stereo platform, the measured depth error of a single point is no higher than 5 mm.
- (iv)
- To prove the effectiveness of the proposed methodology on 3D plant imaging by testing with real greenhouse crops. The proposed methodology generates satisfactory colored point clouds of greenhouse crops in different environments with disparity refinement. It also shows invariance against changing illumination, as well as a capability of recovering 3D surfaces of highlighted leaf regions.
2. Materials
2.1. Stereo Vision Platform
2.2. Sample Plants and Environments
3. Methodology
3.1. Framework
3.2. Calibration
3.3. Stereo Rectification
3.4. Stereo Matching
3.4.1. Raw Matching Cost Computation
3.4.2. Cost Aggregation
3.4.3. Disparity Computation and Disparity Refinement
3.5. 3D Point Cloud~Reconstruction
4. Results
4.1. Performance of the Proposed Stereo Matching Algorithm
4.2. Relationship between Accuracy and Baseline
4.3. Reconstruct Point Cloud with Disparity Refinement
4.4. Implementation Details
5. Discussion
5.1. Depth Error
5.2. Feasibility
5.3. Invariance against Illumination Changes
5.4. Leaves Segmentation
6. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Romeo, J.; Pajares, G.; Montalvo, M.; Guerrero, J.M.; Guijarro, M.; Cruz, J.M.D.L. A new Expert System for Greeness Identification in Agricultural Images. Expert Syst. Appl. 2013, 40, 2275C2286. [Google Scholar] [CrossRef]
- Montalvo, M.; Guerrero, J.M.; Romeo, J.; Emmi, L.; Guijarro, M.; Pajares, G. Automatic expert system for weeds/crops identification in images from maize fields. Expert Syst. Appl. 2013, 40, 75–82. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C.; Jones, D.D.; Hindman, T.W. Intensified fuzzy clusters for classifying plant, soil, and residue regions of interest from color images. Comput. Electron. Agric. 2004, 42, 161–180. [Google Scholar] [CrossRef]
- Bruno, O.M.; Plotze, R.D.O.; Falvo, M.; Castro, M.D. Fractal dimension applied to plant identification. Inform. Sci. 2008, 178, 2722–2733. [Google Scholar] [CrossRef]
- Backes, A.R.; Casanova, D.; Bruno, O.M. Plant Leaf Identification Based On Volumetric Fractal Dimension. Int. J. Pattern Recognit. Artif. Intell. 2011, 23, 1145–1160. [Google Scholar] [CrossRef]
- Neto, J.C.; Meyer, G.E.; Jones, D.D. Individual leaf extractions from young canopy images using GustafsonCKessel clustering and a genetic algorithm. Comput. Electron. Agric. 2006, 51, 66–85. [Google Scholar] [CrossRef]
- Zeng, Q.; Miao, Y.; Liu, C.; Wang, S. Algorithm based on marker-controlled watershed transform for overlapping plant fruit segmentation. Opt. Eng. 2009, 48, 027201. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, F.; Shah, S.G.; Ye, Y.; Mao, H. Use of leaf color images to identify nitrogen and potassium deficient tomatoes. Pattern Recognit. Lett. 2011, 32, 1584–1590. [Google Scholar] [CrossRef]
- Scharr, H.; Minervini, M.; French, A.P.; Klukas, C.; Kramer, D.M.; Liu, X.; Luengo, I.; Pape, J.-M.; Polder, G.; Vukadinovic, D.; et al. Leaf segmentation in plant phenotyping: A collation study. Mach. Vis. Appl. 2015, 27, 585–606. [Google Scholar] [CrossRef]
- Pape, J.M.; Klukas, C. 3-D histogram-based segmentation and leaf detection for rosette plants. In ECCV 2014 Workshops; Springer: Cham, Switzerland, 2015; Volume 8928, pp. 61–74. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Susstrunk, S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Liu, X.; Chen, J.; Kramer, D.M. Multi-leaf alignment from fluorescence plant images. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision (WACV), Steamboat Springs, CO, USA, 24–26 March 2014; pp. 437–444. [Google Scholar]
- Barrow, H.G.; Tenenbaum, J.M.; Bolles, R.C.; Wolf, H.C. Parametric Correspondence and Chamfer Matching: Two New Techniques for Image Matching. In Proceedings of the 5th International Joint Conference on Artificial Intelligence (IJCAI’77), Cambridge, MA, USA, 22–25 August 1977; Volume 2, pp. 659–663. [Google Scholar]
- Fernandez, R.; Montes, H.; Salinas, C.; Sarria, J.; Armada, M. Combination of RGB and Multispectral Imagery for Discrimination of Cabernet Sauvignon Grapevine Elements. Sensors 2013, 13, 7838–7859. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lee, W.S.; Wang, K. Identifying blueberry fruit of different growth stages using natural outdoor color images. Comput. Electron. Agric. 2014, 106, 91–101. [Google Scholar] [CrossRef]
- Sansoni, G.; Trebeschi, M.; Docchio, F. State-of-The-Art and Applications of 3D Imaging Sensors in Industry, Cultural Heritage, Medicine, and Criminal Investigation. Sensors 2009, 9, 568–601. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.V.; Kuhnert, L.; Kuhnert, K.D. Structure overview of vegetation detection. A novel approach for efficient vegetation detection using an active lighting system. Robot. Auton. Syst. 2012, 60, 498–508. [Google Scholar] [CrossRef]
- Alenya, G.; Dellen, B.; Torras, C. 3D modelling of leaves from color and ToF data for robotized plant measuring. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Shanghai, China, 9–13 May 2011; pp. 3408–3414. [Google Scholar]
- Fernández, R.; Salinas, C.; Montes, H.; Sarria, J. Multisensory System for Fruit Harvesting Robots. Experimental Testing in Natural Scenarios and with Different Kinds of Crops. Sensors 2014, 14, 23885–23904. [Google Scholar] [CrossRef] [PubMed]
- Garrido, M.; Paraforos, D.; Reiser, D.; Vzquez Arellano, M.; Griepentrog, H.; Valero, C. 3D Maize Plant Reconstruction Based on Georeferenced Overlapping LiDAR Point Clouds. Remote Sens. 2015, 7, 17077–17096. [Google Scholar] [CrossRef]
- Seidel, D.; Beyer, F.; Hertel, D.; Fleck, S.; Leuschner, C. 3D-laser scanning: A non-destructive method for studying above- ground biomass and growth of juvenile trees. Agric. Forest Meteorol. 2011, 151, 1305–1311. [Google Scholar] [CrossRef]
- Xu, H.; Gossett, N.; Chen, B. Knowledge and heuristic-based modeling of laser-scanned trees. ACM Trans. Graph. 2007, 26, 377–388. [Google Scholar] [CrossRef]
- Dassot, M.; Colin, A.; Santenoise, P.; Fournier, M.; Constant, T. Terrestrial laser scanning for measuring the solid wood volume, including branches, of adult standing trees in the forest environment. Comput. Electron. Agric. 2012, 89, 86–93. [Google Scholar] [CrossRef]
- Dornbusch, T.; Wernecke, P.; Diepenbrock, W. A method to extract morphological traits of plant organs from 3D point clouds as a database for an architectural plant model. Ecol. Modelling 2007, 200, 119–129. [Google Scholar] [CrossRef]
- Li, Y.; Fan, X.; Mitra, N.J.; Chamovitz, D.; Cohen-Or, D.; Chen, B. Analyzing growing plants from 4D point cloud data. ACM Trans. Graph. 2013, 32, 1–10. [Google Scholar] [CrossRef]
- Paulus, S.; Behmann, J.; Mahlein, A.K.; Kuhlmann, H. Low-cost 3D systems: Suitable tools for plant phenotyping. Sensors 2014, 14, 3001–3018. [Google Scholar] [CrossRef] [PubMed]
- Chn, Y.; Rousseau, D.; Lucidarme, P.; Bertheloot, J.; Caffier, V.; Morel, P.; Tienne, B.; Chapeau-Blondeau, F. On the use of depth camera for 3D phenotyping of entire plants. Comput. Electron. Agric. 2012, 82, 122–127. [Google Scholar] [CrossRef]
- Li, D.; Xu, L.; Tan, C.; Goodman, E.D.; Fu, D.; Xin, L. Digitization and visualization of greenhouse tomato plants in indoor environments. Sensors 2015, 15, 4019–4051. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Hayashi, S.; Tsubota, S. Growth measurement of a community of strawberries using three-dimensional sensor. Environ. Control. Biol. 2015, 53, 49–53. [Google Scholar] [CrossRef]
- Schima, R.; Mollenhauer, H.; Grenzdorffer, G.; Merbach, I.; Lausch, A.; Dietrich, P.; Bumberger, J. Imagine all the plants: Evaluation of a light-field camera for on-site crop growth monitoring. Remote Sens. 2016, 8, 823. [Google Scholar] [CrossRef]
- Apelt, F.; Breuer, D.; Nikoloski, Z.; Stitt, M.; Kragler, F. Phytotyping 4D: A light-field imaging system for non-invasive and accurate monitoring of spatio-temporal plant growth. Plant J. 2015, 82, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Biskup, B.; Scharr, H.; Schurr, U.; Rascher, U. A stereo imaging system for measuring structural parameters of plant canopies. Plant Cell Environ. 2007, 30, 1299C1308. [Google Scholar] [CrossRef] [PubMed]
- Teng, C.H.; Kuo, Y.T.; Chen, Y.S. Leaf segmentation, classification, and three-dimensional recovery from a few images with close viewpoints. Opt. Eng. 2011, 50, 103–108. [Google Scholar]
- Hu, P.; Guo, Y.; Li, B.; Zhu, J.; Ma, Y. Three-dimensional reconstruction and its precision evaluation of plant architecture based on multiple view stereo method. Trans. Chin. Soc. Agric. Eng. 2015, 31, 209–214. [Google Scholar]
- Duan, T.; Chapman, S.C.; Holland, E.; Rebetzke, G.J.; Guo, Y.; Zheng, B. Dynamic quantification of canopy structure to characterize early plant vigour in wheat genotypes. J. Exp. Botany 2016, 67, 4523–4534. [Google Scholar] [CrossRef] [PubMed]
- Boykov, Y.; Veksler, O.; Zabih, R. Fast approximate energy minimization via graph cuts. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 1222–1239. [Google Scholar] [CrossRef]
- Hirschmuller, H. Stereo processing by semiglobal matching and mutual information. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 30, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.J.; Kweon, I.S. Adaptive support-weight approach for correspondence search. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Hosni, A.; Rhemann, C.; Bleyer, M.; Rother, C.; Gelautz, M. Fast cost-volume filtering for visual correspondence and beyond. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Hosni, A.; Bleyer, M.; Gelautz, M. Secrets of adaptive support weight techniques for local stereo matching. Comput. Vis. Image Underst. 2013, 117, 620–632. [Google Scholar] [CrossRef]
- Mei, X.; Sun, X.; Zhou, M.; Jiao, S.; Wang, H.; Zhang, X. On building an accurate stereo matching system on graphics hardware. In Proceedings of the IEEE International Conference on Computer Vision Workshops, ICCV 2011 Workshops, Barcelona, Spain, 6–13 November 2011; pp. 467–474. [Google Scholar]
- Bouguet, J.Y. Camera Calibration Toolbox for MATLAB. Available online: http://www.vision.caltech.edu/bouguetj/calib_doc/ (accessed on 14 October 2015).
- Zhang, Z. A Flexible New Technique for Camera Calibration. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1330–1334. [Google Scholar] [CrossRef]
- Tombari, F.; Mattoccia, S.; Stefano, L.D.; Addimanda, E. Classification and evaluation of cost aggregation methods for stereo correspondence. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Anchorage, AK, USA, 23–28 June 2008; pp. 1–8. [Google Scholar]
- Scharstein, D.; Szeliski, R. A Taxonomy and Evaluation of Dense Two-Frame Stereo Correspondence Algorithms. Int. J. Comput. Vis. 2002, 47, 131–140. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, L.; Yang, R.; Stewnius, H.; Nistr, D. Stereo matching with color-weighted correlation, hierarchical belief propagation, and occlusion handling. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 31, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Middlebury Stereo Evaluation-Version 2. Available online: http://vision.middlebury.edu/stereo/eval (accessed on 22 March 2015).
- Chang, C.; Chatterjee, S. Quantization error analysis in stereo vision. In Proceedings of the 1992 Conference Record of The Twenty-Sixth Asilomar Conference on Signals, Systems and Computers, Pacific Grove, CA, USA, 26–28 October 1992; Volume 2, pp. 1037–1041. [Google Scholar]























Tsukuba | Venus | Teddy | Cone | Avg. Error | |
|---|---|---|---|---|---|
| Unit: % | n.o. all dis | n.o. all dis | n.o. all dis | n.o. all dis | |
| Proposed | 2.45 3.53 6.58 | 0.38 1.09 4.39 | 6.97 10.7 18.9 | 2.91 11.6 8.75 | 6.63 |
| ASW [39] | 1.38 1.85 6.90 | 0.71 1.19 6.13 | 7.88 13.3 18.6 | 3.97 9.79 8.26 | 6.67 |
| GC [37] | 1.94 4.12 9.39 | 1.79 3.44 8.75 | 16.5 25.0 24.9 | 7.70 18.2 15.3 | 11.4 |
| SGBM [38] | 4.36 6.47 18.8 | 5.90 7.52 26.3 | 15.5 24.2 26.9 | 12.2 22.1 20.4 | 15.9 |
| Steps: | Image Acquisition | Calibration | Rectification | Stereo Matching | 3D Reconstruction and Display |
|---|---|---|---|---|---|
| Software/Equipment | VS2010+OPENCV2.4.9/The proposed platform | Matlab2014a/Laptop | VS2010+OPENCV2.4.9/Laptop | VS2010+OPENCV2.4.9/Laptop | VS2013+PCL1.7.2 (×86)/Laptop |
| URLs | Microsoft.com; Opencv.org | Vision.caltech.edu/bouguetj/calib_doc/ | Microsoft.com; Opencv.org | Microsoft.com; Opencv.org | Microsoft.com; Pointclouds.org |
| Time (minute) | Less than 3 | About 20 | Less than 1 | Less than 2 | Less than 1 |
| Data size | 40 + 2 images | 40 images | 2 images | 1 scene (point cloud) | 1 scene (point cloud) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Xu, L.; Tang, X.-s.; Sun, S.; Cai, X.; Zhang, P. 3D Imaging of Greenhouse Plants with an Inexpensive Binocular Stereo Vision System. Remote Sens. 2017, 9, 508. https://doi.org/10.3390/rs9050508
Li D, Xu L, Tang X-s, Sun S, Cai X, Zhang P. 3D Imaging of Greenhouse Plants with an Inexpensive Binocular Stereo Vision System. Remote Sensing. 2017; 9(5):508. https://doi.org/10.3390/rs9050508
Chicago/Turabian StyleLi, Dawei, Lihong Xu, Xue-song Tang, Shaoyuan Sun, Xin Cai, and Peng Zhang. 2017. "3D Imaging of Greenhouse Plants with an Inexpensive Binocular Stereo Vision System" Remote Sensing 9, no. 5: 508. https://doi.org/10.3390/rs9050508
APA StyleLi, D., Xu, L., Tang, X. -s., Sun, S., Cai, X., & Zhang, P. (2017). 3D Imaging of Greenhouse Plants with an Inexpensive Binocular Stereo Vision System. Remote Sensing, 9(5), 508. https://doi.org/10.3390/rs9050508