Chemiluminescent Receptor Binding Assay for Ciguatoxins and Brevetoxins Using Acridinium Brevetoxin-B2
Abstract
:1. Introduction
2. Results
2.1. Preparation of ABTX
2.2. Affinity of ABTX to Rat Brain Synaptosomes
2.3. Saturation Experiment to Rat Synaptosome by ABTX
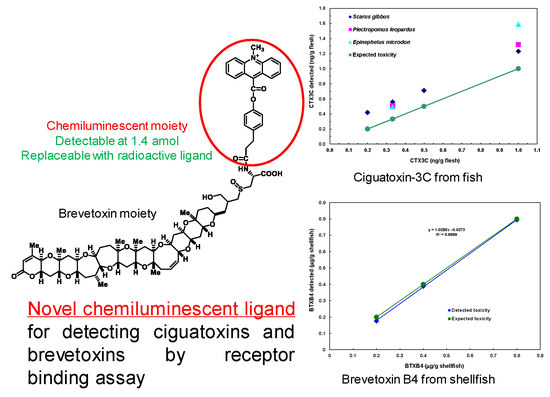
2.4. Binding Assay Using ABTX for Detection of CTX3C and BTXB2
2.5. Detection of CTX3C from Fish
2.6. Detection of BTXB4 from Mussels
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of ABTX
4.3. Measurement of Chemiluminescent Intensity
4.4. Preparation of a Synaptosome Fraction from Rat Brain
4.5. Binding Assay Using 3H-PbTx-3 for Evaluating the Affinity of ABTX against Rat Brain Synaptosome
4.6. Binding Assay Using ABTX for the Detection of CTX or BTX
4.7. Assay of CTX3C in Fish Flesh Using ABTX
4.8. Assay of BTXB4 in Blue Mussels Using ABTX
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yasumoto, T.; Satake, M. Chemistry, Etiology and Determination Methods of Ciguatera Toxins. J. Toxicol. Toxin Rev. 1996, 15, 91–107. [Google Scholar] [CrossRef]
- Yasumoto, T.; Nakajima, I.; Bagnis, R.; Adachi, R. Finding of a dinoflagellate as a likely culprit of ciguatera. Nippon. SUISAN GAKKAISHI 1977, 43, 1021–1026. [Google Scholar] [CrossRef]
- Murata, M.; Legrand, A.M.; Ishibashi, Y.; Fukui, M.; Yasumoto, T. Structures and configurations of ciguatoxin from the moray eel Gymnothorax javanicus and its likely precursor from the dinoflagellate Gambierdiscus toxicus. J. Am. Chem. Soc. 1990, 112, 4380–4386. [Google Scholar] [CrossRef]
- Yasumoto, T. The chemistry and biological function of natural marine toxins. Chem. Rec. 2001, 1, 228–242. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, T.; Kuniyoshi, K.; Oshiro, N.; Yasumoto, T. Biooxidation of Ciguatoxins Leads to Species-Specific Toxin Profiles. Toxins 2017, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, B.; Hurbungs, M.; Jones, A.; Lewis, R.J. Multiple ciguatoxins present in Indian Ocean reef fish. Toxicon 2002, 40, 1347–1353. [Google Scholar] [CrossRef]
- Abraham, A.; Jester, E.L.; Granade, H.R.; Plakas, S.M.; Dickey, R.W. Caribbean ciguatoxin profile in raw and cooked fish implicated in ciguatera. Food Chem. 2012, 131, 192–198. [Google Scholar] [CrossRef]
- Tsumuraya, T.; Fujii, I.; Hirama, M. Production of monoclonal antibodies for sandwich immunoassay detection of Pacific ciguatoxins. Toxicon 2010, 56, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Nozawa, A.; Totoribe, K.; Muramatsu, N.; Nukaya, H.; Tsuji, K.; Yamaguchi, K.; Yasumoto, T.; Kaspar, H.; Berkett, N.; et al. Brevetoxin B1, a new polyether marine toxin from the New Zealand shellfish, Austrovenus stutchburyi. Tetrahedron Lett. 1995, 36, 725–728. [Google Scholar] [CrossRef]
- Morohashi, A.; Satake, M.; Murata, K.; Naoki, H.; Kaspar, H.F.; Yasumoto, T. Brevetoxin B3, a new brevetoxin analog isolated from the greenshell mussel perna canaliculus involved in neurotoxic shellfish poisoning in new zealand. Tetrahedron Lett. 1995, 36, 8995–8998. [Google Scholar] [CrossRef]
- Murata, K.; Satake, M.; Naoki, H.; Kaspar, H.F.; Yasumoto, T. Isolation and structure of a new brevetoxin analog, brevetoxin B2, from greenshell mussels from New Zealand. Tetrahedron 1998, 54, 735–742. [Google Scholar] [CrossRef]
- Morohashi, A.; Satake, M.; Naoki, H.; Kaspar, H.F.; Oshima, Y.; Yasumoto, T. Brevetoxin B4 isolated from greenshell mussels Perna canaliculus, the major toxin involved in neurotoxic shellfish poisoning in New Zealand. Nat. Toxins 1999, 7, 45–48. [Google Scholar] [CrossRef]
- Ishida, H.; Nozawa, A.; Hamano, H.; Naoki, H.; Fujita, T.; Kaspar, H.F.; Tsuji, K. Brevetoxin B5, a new brevetoxin analog isolated from cockle Austrovenus stutchburyi in New Zealand, the marker for monitoring shellfish neurotoxicity. Tetrahedron Lett. 2004, 45, 29–33. [Google Scholar] [CrossRef]
- Abraham, A.; Plakas, S.M.; Flewelling, L.J.; El Said, K.R.; Jester, E.L.; Granade, H.R.; White, K.D.; Dickey, R.W. Biomarkers of Neurotoxic Shellfish Poisoning. Toxicon 2008, 52, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Satake, M.; Murata, M.; Yasumoto, T. The structure of CTX3C, a ciguatoxin congener isolated from cultured Gambierdiscus toxicus. Tetrahedron Lett. 1993, 34, 1975–1978. [Google Scholar] [CrossRef]
- Whitney, P.L.; Delgado, J.A.; Baden, D.G. Complex behavior of marine animal tissue extracts in the competitive binding assay of brevetoxins with rat brain synaptosomes. Nat. Toxins 1997, 5, 193–200. [Google Scholar] [CrossRef] [PubMed]
- McCall, J.R.; Jacocks, H.M.; Baden, D.G.; Bourdelais, A.J. Development of a competitive fluorescence-based synaptosome binding assay for brevetoxins. Harmful Algae 2012, 19, 85–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardison, D.R.; Holland, W.C.; McCall, J.R.; Bourdelais, A.J.; Baden, D.G.; Darius, H.T.; Chinain, M.; Tester, P.A.; Shea, D.; Quintana, H.A.F.; et al. Fluorescent Receptor Binding Assay for Detecting Ciguatoxins in Fish. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Weeks, I.; Beheshti, I.; Mccapra, F.; Campbell, A.K.; Woodhead, J.S. Acridinium esters as high-specific-activity labels in immunoassay. Clin. Chem. 1983, 29, 1474–1479. [Google Scholar] [PubMed]
- A Poli, M.; Mende, T.J.; Baden, D.G. Brevetoxins, unique activators of voltage-sensitive sodium channels, bind to specific sites in rat brain synaptosomes. Mol. Pharmacol. 1986, 30, 129–135. [Google Scholar] [PubMed]











© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murata, K.; Yasumoto, T. Chemiluminescent Receptor Binding Assay for Ciguatoxins and Brevetoxins Using Acridinium Brevetoxin-B2. Toxins 2019, 11, 580. https://doi.org/10.3390/toxins11100580
Murata K, Yasumoto T. Chemiluminescent Receptor Binding Assay for Ciguatoxins and Brevetoxins Using Acridinium Brevetoxin-B2. Toxins. 2019; 11(10):580. https://doi.org/10.3390/toxins11100580
Chicago/Turabian StyleMurata, Kazuya, and Takeshi Yasumoto. 2019. "Chemiluminescent Receptor Binding Assay for Ciguatoxins and Brevetoxins Using Acridinium Brevetoxin-B2" Toxins 11, no. 10: 580. https://doi.org/10.3390/toxins11100580
APA StyleMurata, K., & Yasumoto, T. (2019). Chemiluminescent Receptor Binding Assay for Ciguatoxins and Brevetoxins Using Acridinium Brevetoxin-B2. Toxins, 11(10), 580. https://doi.org/10.3390/toxins11100580