AAU-Specific RNA Cleavage Mediated by MazF Toxin Endoribonuclease Conserved in Nitrosomonas europaea
Abstract
:1. Introduction
2. Results
2.1. Enzymatic Activity of MazFNE1181
2.2. Cleavage Sequence Identification Using Massive Parallel Sequencing
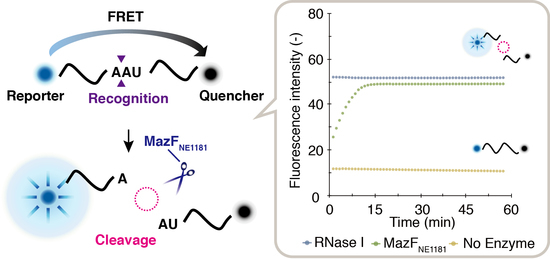
2.3. Cleavage-Specificity Validation Based on Fluorescence Resonance Energy Transfer
3. Discussion
4. Materials and Methods
4.1. Plasmids and Oligonucleotides
4.2. Plasmid Construction
4.3. Expression of MazENE1182
4.4. Purification of MazENE1182
4.5. Expression of MazFNE1181
4.6. Purification of MazFNE1181
4.7. Enzymatic Activity of MazFNE1181 and MazENE1182
4.8. Endoribonuclease Activity of MazFNE1181
4.9. Cleavage Sequence Identification
4.10. Fluorometric Detection of MazFNE1181 Activity
4.11. Neutralization of MazFNE1181-mediated Cleavage
4.12. Accession Numbers
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| TA | toxin-antitoxin |
| 6-FAM | 6-carboxyfluorescein |
| BHQ-1 | black hole quencher-1 |
| RASTA-Bacteria | rapid automated scan for toxins and antitoxins in bacteria |
Appendix A
Appendix B
References
- Arp, D.J.; Sayavedra-Soto, L.A.; Hommes, N.G. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 2002, 178, 250–255. [Google Scholar]
- Bothe, H.; Jost, G.; Schloter, M.; Ward, B.B.; Witzel, K. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 2000, 24, 673–690. [Google Scholar] [CrossRef] [PubMed]
- Arp, D.J.; Stein, L.Y. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 471–495. [Google Scholar] [CrossRef] [PubMed]
- Groeneweg, J.; Sellner, B.; Tappe, W. Ammonia oxidation in Nitrosomonas at NH3 concentrations near km: Effects of pH and temperature. Water Res. 1994, 28, 2561–2566. [Google Scholar] [CrossRef]
- Stein, L.Y.; Arp, D.J. Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol. 1998, 64, 4098–4102. [Google Scholar] [PubMed]
- Park, S.; Ely, R.L. Candidate stress genes of Nitrosomonas europaea for monitoring inhibition of nitrification by heavy metals. Appl. Environ. Microbiol. 2008, 74, 5475–5482. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ely, R.L. Genome-wide transcriptional responses of Nitrosomonas europaea to zinc. Arch. Microbiol. 2008, 189, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Gvakharia, B.O.; Permina, E.A.; Gelfand, M.S.; Bottomley, P.J.; Sayavedra-Soto, L.A.; Arp, D.J. Global transcriptional response of Nitrosomonas europaea to chloroform and chloromethane. Appl. Environ. Microbiol. 2007, 73, 3440–3445. [Google Scholar] [CrossRef] [PubMed]
- Chain, P.; Lamerdin, J.; Larimer, F.; Lao, V.; Land, M.; Hauser, L.; Klotz, M.; Norton, J.; Arciero, D.; Hommes, N.; et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 2003, 185, 2759–2773. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Bertram, R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 2013, 340, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Inouye, M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011, 9, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Aizenman, E.; Engelberg-Kulka, H.; Glaser, G. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3’,5'-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 1996, 93, 6059–6063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Hoeflich, K.P.; Ikura, M.; Qing, G.; Inouye, M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 2003, 12, 913–923. [Google Scholar] [CrossRef]
- Amitai, S.; Kolodkin-Gal, I.; Hananya-Meltabashi, M.; Sacher, A.; Kulka, H.E. Escherichia coli MazF leads to the simultaneous selective synthesis of both "death proteins" and "survival proteins". PLoS Genet. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Vesper, O.; Amitai, S.; Belitsky, M.; Byrgazov, K.; Kaberdina, A.C.; Engelberg-Kulka, H.; Moll, I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 2011, 147, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Chopra, N.; Saumitra; Pathak, A.; Bhatnagar, R.; Bhatnagar, S. Linkage, mobility, and selfishness in the MazF family of bacterial toxins: A snapshot of bacterial evolution. Genome Biol. Evol. 2013, 5, 2268–2284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, Y.; Zhu, L.; Suzuki, M.; Inouye, M. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 2004, 279, 20678–20684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, L.; Zhang, J.; Inouye, M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 2005, 280, 26080–26088. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Yamaguchi, Y.; Inouye, M. Bacillus subtilis MazF-bs (EndoA) is a UACAU-specific mRNA interferase. FEBS Lett. 2011, 585, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Inoue, K.; Yoshizumi, S.; Kobayashi, H.; Zhang, Y.; Ouyang, M.; Kato, F.; Sugai, M.; Inouye, M. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 2009, 191, 3248–3255. [Google Scholar] [CrossRef] [PubMed]
- Rothenbacher, F.P.; Suzuki, M.; Hurley, J.M.; Montville, T.J.; Kirn, T.J.; Ouyang, M.; Woychik, N.A. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 2012, 194, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Nariya, H.; Park, J.-H.; Inouye, M. Inhibition of specific gene expressions by protein-mediated mRNA interference. Nat. Commun. 2012, 3, 607. [Google Scholar] [CrossRef] [PubMed]
- Syed, M.A.; Koyanagi, S.; Sharma, E.; Jobin, M.C.; Yakunin, A.F.; Lévesque, C.M. The chromosomal mazEF locus of Streptococcus mutans encodes a functional type II toxin-antitoxin addiction system. J. Bacteriol. 2011, 193, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Kato, Y.; Sekiguchi, Y.; Tsuneda, S.; Noda, N. Characterization of MazF-mediated sequence-specific RNA cleavage in Pseudomonas putida using massive parallel sequencing. PLoS ONE 2016, 11, e0149494. [Google Scholar] [CrossRef] [PubMed]
- Bukowski, M.; Lyzen, R.; Helbin, W.M.; Bonar, E.; Szalewska-Palasz, A.; Wegrzyn, G.; Dubin, G.; Dubin, A.; Wladyka, B. A regulatory role for Staphylococcus aureus toxin-antitoxin system PemIKSa. Nat. Commun. 2013, 4, 2012. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Phadtare, S.; Nariya, H.; Ouyang, M.; Husson, R.N.; Inouye, M. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 2008, 69, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, Y.; Teh, J.S.; Zhang, J.; Connell, N.; Rubin, H.; Inouye, M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 18638–18643. [Google Scholar] [CrossRef] [PubMed]
- Nariya, H.; Inouye, M. MazF, an mRNA Interferase, Mediates Programmed Cell Death during Multicellular Myxococcus Development. Cell 2008, 132, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Dewan, P.C.; Siddique, S.A.; Varadarajan, R. MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 2014, 289, 4191–4205. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 2004, 272, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Alawneh, A.M.; Qi, D.; Yonesaki, T.; Otsuka, Y. An ADP-ribosyltransferase Alt of bacteriophage T4 negatively regulates the Escherichia coli MazF toxin of a toxin-antitoxin module. Mol. Microbiol. 2015, 99, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Arora, G.; Singh, M.; Kidwai, S.; Narayan, O.P.; Singh, R. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 2015, 6, 6059. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.R.; Hergenrother, P.J. A continuous fluorometric assay for the assessment of MazF ribonuclease activity. Anal. Biochem. 2007, 371, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 2009, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Short, F.L.; Rao, F.; Mizuguchi, K.; Pei, X.Y.; Fineran, P.C.; Luisi, B.F.; Salmond, G.P.C. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012, 40, 6158–6173. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.K.; Gerdes, K. RelE toxins from Bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 2003, 48, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Prysak, M.H.; Mozdzierz, C.J.; Cook, A.M.; Zhu, L.; Zhang, Y.; Inouye, M.; Woychik, N.A. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009, 71, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Christensen-Dalsgaard, M.; Jørgensen, M.G.; Gerdes, K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010, 75, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, M.G.; Pandey, D.P.; Jaskolska, M.; Gerdes, K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 2009, 191, 1191–1199. [Google Scholar] [CrossRef] [PubMed]
- Arcus, V.L.; Mckenzie, J.L.; Robson, J.; Cook, G.M. The PIN-domain ribonucleases and the prokaryotic VapBC toxin-antitoxin array. Protein Eng. Des. Sel. 2011, 24, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Takayama, M.; Asada, K.; Kato, I. Endoribonuclease. U.S. Patent 7,989,184, 5 June 2012. [Google Scholar]
- Schifano, J.M.; Vvedenskaya, I.O.; Knoblauch, J.G.; Ouyang, M.; Nickels, B.E.; Woychik, N.A. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014, 5, 3538. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Hong, S.H.; Peti, W.; Benedik, M.J.; Page, R.; Wood, T.K. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 2013, 15, 1734–1744. [Google Scholar] [CrossRef] [PubMed]
- Sevin, E.W.; Barloy-Hubler, F. RASTA-Bacteria: A web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007, 8, R155. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Christensen, S.K.; Løbner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.; Hon, G.; Chandonia, J.; Brenner, S. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]



| Name | Sequence (5′ to 3′) a |
|---|---|
| DR-13-AAU | AAAAAAAUAAAAA |
| DR-13-AAA | AAAAAAAAAAAAA |
| D-13-AAA | AAAAAAAAAAAAA |
| R-13-GUUGU | GUUGUCAUGCCGG |
| R-13-UCUCG | UCUCGGUGCGUUG |
| R-13-UGACA | UGACACGAACCGC |
| DR-13-GAU | AAAAAGAUAAAAA |
| DR-13-AAC | AAAAAAACAAAAA |
| Locus | Gene Symbol | Length (bp) | Product Name |
|---|---|---|---|
| NE0390 | rpmH | 135 | LSU Ribosomal protein L34 |
| NE2575 | merE | 237 | mercury resistance protein |
| NE0841 | merP | 276 | mercury scavenger protein |
| NE0842 | merT | 351 | mercuric transport protein |
| NE1224 | - | 264 | hypothetical protein |
| NE1344 | - | 279 | hypothetical protein |
| NE2523 | - | 231 | hypothetical protein |
| NE2538 | - | 912 | hypothetical protein |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miyamoto, T.; Yokota, A.; Tsuneda, S.; Noda, N. AAU-Specific RNA Cleavage Mediated by MazF Toxin Endoribonuclease Conserved in Nitrosomonas europaea. Toxins 2016, 8, 174. https://doi.org/10.3390/toxins8060174
Miyamoto T, Yokota A, Tsuneda S, Noda N. AAU-Specific RNA Cleavage Mediated by MazF Toxin Endoribonuclease Conserved in Nitrosomonas europaea. Toxins. 2016; 8(6):174. https://doi.org/10.3390/toxins8060174
Chicago/Turabian StyleMiyamoto, Tatsuki, Akiko Yokota, Satoshi Tsuneda, and Naohiro Noda. 2016. "AAU-Specific RNA Cleavage Mediated by MazF Toxin Endoribonuclease Conserved in Nitrosomonas europaea" Toxins 8, no. 6: 174. https://doi.org/10.3390/toxins8060174
APA StyleMiyamoto, T., Yokota, A., Tsuneda, S., & Noda, N. (2016). AAU-Specific RNA Cleavage Mediated by MazF Toxin Endoribonuclease Conserved in Nitrosomonas europaea. Toxins, 8(6), 174. https://doi.org/10.3390/toxins8060174