Type II Toxin–Antitoxin Systems in the Unicellular Cyanobacterium Synechocystis sp. PCC 6803
Abstract
:1. Introduction
2. Results
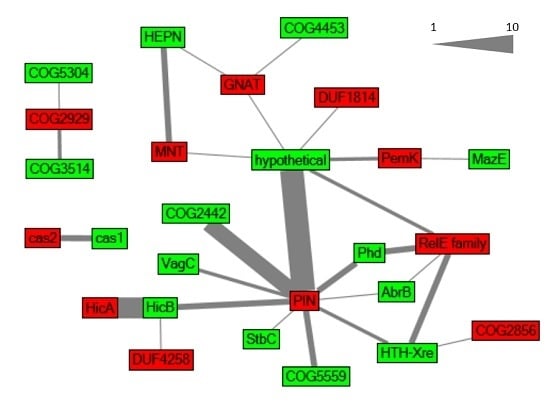
2.1. Global Characterization of Synechocystis 6803 Type II TA Systems
2.2. Verification of Predicted TA Functionality
2.3. Synechocystis 6803 Type II TA Systems of Special Interest
2.4. Synechocystis 6803 Type II TA Systems in the Maintenance of Plasmids and Genomic Islands
2.4.1. The Slr6057–Slr6056 TA System May Stabilize a Genetic Cassette for a Bacterial Multiubiquitin System on Plasmid pSYSX
2.4.2. Is the hoxEFUYH Hydrogenase Operon in Synechocystis 6803 Stabilized by a TA System?
2.4.3. Two Type II TA Systems Are Part of the Large 40 kb Genomic Island in Synechocystis 6803
2.5. RNase Activity Is the Most Frequent Characteristics Associated with Type II TA Systems in Synechocystis 6803
2.5.1. The RelE Protein Ssl7039 Is a Ribosome-Independent Ribonuclease in Vitro and Leads to the Degradation of E. coli lpp mRNA in Vivo
2.5.2. Heterologous Expression of Slr8014 in E. coli Causes Degradation of lpp mRNA in Vivo That Is Abolished by the Associated Antitoxin
3. Discussion
3.1. Transcriptional Features of Synechocystis 6803 Type II TA Systems
3.2. Similarities of Synechocystis 6803 Type II TA Systems to Those of Other Cyanobacteria
3.3. Synechocystis 6803 Type II TA Systems with a Putative Function in the Maintenance of Plasmids and Genomic Islands
4. Conclusions
5. Materials and Methods
5.1. Comprehensive TA System Detection
5.2. Experimental Methods
5.2.1. Growth Inhibition Assay
5.2.2. Overexpression and Purification of Toxins and Antitoxins
5.2.3. In Vitro Synthesis of RNA
5.2.4. RNA Extraction
5.2.5. RNase Activity
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CRISPR | clustered regularly interspaced short palindromic repeats |
| DUF | domain of unknown function |
| TA | toxin–antitoxin |
| TADB | toxin–antitoxin database |
| TSS | transcriptional start site |
| TU | transcriptional unit |
References
- Schuster, C.F.; Bertram, R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 2013, 340, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.T.; Espinosa, M.; Yeo, C.C. Keeping the Wolves at Bay: Antitoxins of Prokaryotic Type II Toxin-Antitoxin Systems. Front. Mol. Biosci. 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Leplae, R.; Geeraerts, D.; Hallez, R.; Guglielmini, J.; Drèze, P.; Van Melderen, L. Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011, 39, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Rocker, A.; Meinhart, A. Type II toxin: Antitoxin systems. More than small selfish entities? Curr. Genet. 2016, 62, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.F.; Bertram, R. Toxin-Antitoxin Systems of Staphylococcus aureus. Toxins 2016, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Park, J.-H.; Inouye, M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011, 45, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Short, F.L.; Rao, F.; Mizuguchi, K.; Pei, X.Y.; Fineran, P.C.; Luisi, B.F.; Salmond, G.P.C. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 2012, 40, 6158–6173. [Google Scholar] [CrossRef] [PubMed]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of type I toxin-antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S.; Jahn, N. sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 2015, 39, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Markovski, M.; Wickner, S. Preventing bacterial suicide: A novel toxin-antitoxin strategy. Mol. Cell 2013, 52, 611–612. [Google Scholar] [CrossRef] [PubMed]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A Bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 2013, 52, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Shaw, K.J. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 2003, 185, 6600–6608. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lord, D.M.; Cheng, H.-Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.-Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39. [Google Scholar] [CrossRef] [PubMed]
- TADB. Available online: http://202.120.12.135/TADB2/index.php (accessed on 8 June 2016).
- Ning, D.; Ye, S.; Liu, B.; Chang, J. The proteolytic activation of the relNEs (ssr1114/slr0664) toxin-antitoxin system by both proteases Lons and ClpP2s/Xs of Synechocystis sp. PCC 6803. Curr. Microbiol. 2011, 63, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.; Zhao, W.; Qian, Y. A hypothetical gene pair, ssl2138 and sll11092, constitutes a functional TA system on the chromosome of Synechocystis PCC 6803. Wei Sheng Wu Xue Bao 2013, 53, 1043–1049. [Google Scholar] [PubMed]
- Ning, D.; Liu, S.; Xu, W.; Zhuang, Q.; Wen, C.; Tang, X. Transcriptional and proteolytic regulation of the toxin-antitoxin locus vapBC10 (ssr2962/slr1767) on the chromosome of Synechocystis sp. PCC 6803. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Krishna, P.S.; Rani, B.R.; Mohan, M.K.; Suzuki, I.; Shivaji, S.; Prakash, J.S.S. A novel transcriptional regulator, Sll1130, negatively regulates heat-responsive genes in Synechocystis sp. PCC6803. Biochem. J. 2013, 449, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Čelešnik, H.; Tanšek, A.; Tahirović, A.; Vižintin, A.; Mustar, J.; Vidmar, V.; Dolinar, M. Biosafety of biotechnologically important microalgae: Intrinsic suicide switch implementation in cyanobacterium Synechocystis sp. PCC 6803. Biol. Open 2016, 5, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Kopfmann, S.; Hess, W.R. Toxin-antitoxin systems on the large defense plasmid pSYSA of Synechocystis sp. PCC 6803. J. Biol. Chem. 2013, 288, 7399–7409. [Google Scholar] [CrossRef] [PubMed]
- Sevin, E.W.; Barloy-Hubler, F. RASTA-Bacteria: A web-based tool for identifying toxin-antitoxin loci in prokaryotes. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Blower, T.R.; Salmond, G.P.C.; Luisi, B.F. Balancing at survival’s edge: The structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Curr. Opin. Struct. Biol. 2011, 21, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Anantharaman, V.; Aravind, L.; Koonin, E.V. Live virus-free or die: Coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, J.; Georg, J.; Scholz, I.; Sharma, C.M.; Dienst, D.; Bantscheff, J.; Voss, B.; Steglich, C.; Wilde, A.; Vogel, J.; Hess, W.R. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 2011, 108, 2124–2129. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Klähn, S.; Scholz, I.; Matthiessen, J.K.F.; Hess, W.R.; Voß, B. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014, 21, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, J.K.; Lee, A.S.Y.; Engelman, A.; Doudna, J.A. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 2015, 519, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, J.K.; Kranzusch, P.J.; Noeske, J.; Wright, A.V.; Davies, C.W.; Doudna, J.A. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014, 21, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, D.; Voß, B.; Wilde, A.; Al-Babili, S.; Hess, W.R. Microevolution in cyanobacteria: Re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 2012, 19, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Scholz, I.; Lange, S.J.; Hein, S.; Hess, W.R.; Backofen, R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Sesto, N.; Wurtzel, O.; Archambaud, C.; Sorek, R.; Cossart, P. The excludon: A new concept in bacterial antisense RNA-mediated gene regulation. Nat. Rev. Microbiol. 2013, 11, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Rocker, A.; Meinhart, A. A cis-acting antitoxin domain within the chromosomal toxin-antitoxin module EzeT of Escherichia coli quenches toxin activity. Mol. Microbiol. 2015, 97, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.; Engelberg-Kulka, H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genom. MGG 2004, 272, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P.C. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Dy, R.L.; Przybilski, R.; Semeijn, K.; Salmond, G.P.C.; Fineran, P.C. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 2014, 42, 4590–4605. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Snir, S.; Koonin, E.V. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 2011, 193, 6039–6056. [Google Scholar] [CrossRef] [PubMed]
- Shigi, N. Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J. Biol. Chem. 2012, 287, 17568–17577. [Google Scholar] [CrossRef] [PubMed]
- Humbard, M.A.; Miranda, H.V.; Lim, J.-M.; Krause, D.J.; Pritz, J.R.; Zhou, G.; Chen, S.; Wells, L.; Maupin-Furlow, J.A. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 2010, 463, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Burroughs, A.M.; Aravind, L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Appel, J. The Physiology and Functional Genomics of Cyanobacterial Hydrogenases and Approaches towards Biohydrogen Production. In Functional Genomics and Evolution of Photosynthetic Systems; Burnap, R., Vermaas, W., Eds.; Advances in Photosynthesis and Respiration; Springer Netherlands: Berlin, Germany, 2012; pp. 357–381. [Google Scholar]
- Kopf, M.; Klähn, S.; Pade, N.; Weingärtner, C.; Hagemann, M.; Voß, B.; Hess, W.R. Comparative genome analysis of the closely related Synechocystis strains PCC 6714 and PCC 6803. DNA Res. 2014, 21, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Okamoto, S.; Katayama, T.; Nakao, M.; Yoshimura, H.; Kajiya-Kanegae, H.; Yamamoto, S.; Yano, C.; Yanaka, Y.; Maita, H.; et al. CyanoBase and RhizoBase: Databases of manually curated annotations for cyanobacterial and rhizobial genomes. Nucleic Acids Res. 2014, 42, D666–D670. [Google Scholar] [CrossRef] [PubMed]
- Eckert, C.; Boehm, M.; Carrieri, D.; Yu, J.; Dubini, A.; Nixon, P.J.; Maness, P.-C. Genetic analysis of the Hox hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 reveals subunit roles in association, assembly, maturation, and function. J. Biol. Chem. 2012, 287, 43502–43515. [Google Scholar] [CrossRef] [PubMed]
- Aubert-Jousset, E.; Cano, M.; Guedeney, G.; Richaud, P.; Cournac, L. Role of HoxE subunit in Synechocystis PCC6803 hydrogenase. FEBS J. 2011, 278, 4035–4043. [Google Scholar] [CrossRef] [PubMed]
- Jurėnaitė, M.; Markuckas, A.; Sužiedėlienė, E. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 2013, 195, 3165–3172. [Google Scholar] [CrossRef] [PubMed]
- Prysak, M.H.; Mozdzierz, C.J.; Cook, A.M.; Zhu, L.; Zhang, Y.; Inouye, M.; Woychik, N.A. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009, 71, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Rothenbacher, F.P.; Suzuki, M.; Hurley, J.M.; Montville, T.J.; Kirn, T.J.; Ouyang, M.; Woychik, N.A. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 2012, 194, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Sharp, J.D.; Cruz, J.W.; Raman, S.; Inouye, M.; Husson, R.N.; Woychik, N.A. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 2012, 287, 12835–12847. [Google Scholar] [CrossRef] [PubMed]
- Vesper, O.; Amitai, S.; Belitsky, M.; Byrgazov, K.; Kaberdina, A.C.; Engelberg-Kulka, H.; Moll, I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 2011, 147, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.; Moll, I.; Giuliodori, A.M.; Gualerzi, C.O.; Bläsi, U. Temperature-dependent translation of leaderless and canonical mRNAs in Escherichia coli. FEMS Microbiol. Lett. 2002, 211, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Georg, J.; Voß, B.; Scholz, I.; Mitschke, J.; Wilde, A.; Hess, W.R. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Guérout, A.-M.; Krin, E.; Le Roux, F.; Mazel, D. Comprehensive functional analysis of the 18 Vibrio cholerae N16961 toxin-antitoxin systems substantiates their role in stabilizing the superintegron. J. Bacteriol. 2015, 197, 2150–2159. [Google Scholar] [CrossRef] [PubMed]
- Magnuson, R.D. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 2007, 189, 6089–6092. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.P.; Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005, 33, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Averina, O.V.; Alekseeva, M.G.; Abilev, S.K.; Il’in, V.K.; Danilenko, V.N. Distribution of genes of toxin-antitoxin systems of mazEF and relBE families in bifidobacteria from human intestinal microbiota. Genetika 2013, 49, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Klähn, S.; Voss, B.; Stüber, K.; Huettel, B.; Reinhardt, R.; Hess, W.R. Finished genome sequence of the unicellular cyanobacterium Synechocystis sp. strain PCC 6714. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; de Marsac, N.T.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Martins, P.M.M.; Machado, M.A.; Silva, N.V.; Takita, M.A.; de Souza, A.A. Type II Toxin-Antitoxin distribution and adaptive aspects on Xanthomonas genomes: Focus on Xanthomonas citri. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sühnel, J. The Xanthomonas Genome Browser. Available online: http://xgb.fli-leibniz.de/cgi/index.pl (accessed on 14 June 2016).
- Guglielmini, J.; Van Melderen, L. Bacterial toxin-antitoxin systems: Translation inhibitors everywhere. Mob. Genet. Elem. 2011, 1, 283–306. [Google Scholar] [CrossRef] [PubMed]
- Holberger, L.E.; Garza-Sánchez, F.; Lamoureux, J.; Low, D.A.; Hayes, C.S. A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett. 2012, 586, 132–136. [Google Scholar] [CrossRef] [PubMed]
- McNeil, M.B.; Iglesias-Cans, M.C.; Clulow, J.S.; Fineran, P.C. YgfX (CptA) is a multimeric membrane protein that interacts with the succinate dehydrogenase assembly factor SdhE (YgfY). Microbiology 2013, 159, 1352–1365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Inouye, M. RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol. Microbiol. 2011, 79, 1418–1429. [Google Scholar] [CrossRef] [PubMed]
- Kopf, M.; Klähn, S.; Scholz, I.; Hess, W.R.; Voß, B. Variations in the non-coding transcriptome as a driver of inter-strain divergence and physiological adaptation in bacteria. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Scholz, I.; Voß, B.; Hess, W.R. Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 2013, 10, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Pinto, F.; Thapper, A.; Sontheim, W.; Lindblad, P. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Stazic, D.; Lindell, D.; Steglich, C. Antisense RNA protects mRNA from RNase E degradation by RNA–RNA duplex formation during phage infection. Nucleic Acids Res. 2011, 39, 4890–4899. [Google Scholar] [CrossRef] [PubMed]







| Gene ID | Annotation | Re-Annotation |
|---|---|---|
| slr0725 | Chr:110808..111224 | Chr:110727..111224 |
| ssr0756 | Chr:2084575..2084760 | Chr:2084455..2084760 |
| ssr0761 | Chr:2085826..2086110 | Chr:2085859..2086110 |
| ssl1004 | Chr:3262471..3262749 | Chr:3262471..3262707 |
| sll1714 | Chr:968893..969294 | Chr:968893..969132 |
| sll7006 | pSYSA:3367..3591 | pSYSA:3367..3768 |
| ssl0350a | Chr:2738918..2739133 | novel ORF |
| sll0624a | Chr:3341880..3342128 | novel ORF |
| ssr0757a | Chr:2085211..2085417 | novel ORF |
| ssr0761a | Chr:2086107..2086211 | novel ORF |
| slr1062a | Chr:365077..365283 | novel ORF |
| ssl1493a | Chr:529033..529416 | novel ORF |
| ssr2784a | Chr:3474807..3474998 | novel ORF |
| Toxin | Type | A.toxin | Type | TSS | Dis. | Repl. | Ord. | Reference | Comment |
|---|---|---|---|---|---|---|---|---|---|
| sll0205 | PIN | ssl0385 | COG5559/DUF2281 | 2516776 R | 27 | Chr | AT-T | TW | - |
| ssl0258 | HicA (YcfA) | ssl0259 | HicB (COG1598) | 2193884 R | 72 | Chr | T-AT | TW | only second component exhibits TSS |
| sll0286 | GNAT | sll0284 | HEPN (DUF86) | 3024345 R | 74 | Chr | AT-T | TW | - |
| ssr0335 | HicA (YcfA) | ssr0336 | HicB (COG1598) | 2754240 F | 401 | Chr | T-AT | TW | - |
| ssl0350a | HicA (YcfA) | ssl0350 | HicB (COG1598) | 2740742 R | 1609 | Chr | AT-T | TW | TSS of TU |
| sll0406 | PIN | sll0405 | hypothetical | 2552996 R | 34 | Chr | AT-T | TW | potential asRNA regulation |
| sll0525 | PIN | ssl1004 | PhdYeFM_antitox | 3262737 | 30 | Chr | AT-T | [3,16] | see Figure 3 |
| sll0624 | PIN3 | sll0624a | HicB (COG1598) | 3342723 R | 281 | Chr | T-AT | TW | both components exhibit TSS |
| sll0658 | PIN | ssl1255 | HTH (COG2886) | 3121956 R | 3 | Chr | AT-T | [3,16] | - |
| slr0664 | RelE (DUF1044) | ssr1114 | hypothetical | 3347861 F | 80 | Chr | AT-T | [16,18] | - |
| sll0690 | PIN | ssl1300 | VagC | 2632007 R | 139 | Chr | AT-T | [3,16] | alternative TSS (2631867 R) |
| slr0725 | YhaV | slr0724 | MazE (AbrB) | 110366 F | 26 | Chr | AT-T | [3,16] | - |
| ssl0739 | HicA (YcfA) | ssl0738 | HicB (COG1598) | N/K | N/K | Chr | AT-T | TW | - |
| ssr0756 | PIN | ssr0755 | hypothetical | 2084115 F | 28 | Chr | AT-T | TW | - |
| ssr0757 | DUF497/COG2929 | ssr0757a | DUF4415/COG3514 | 2084877 F | 45 | Chr | T-AT | TW | - |
| ssr0761a | YoeB | ssr0761 | PhdYeFM_antitox | 2085833 F | 27 | Chr | AT-T | TW | - |
| slr0771 | PIN | slr0770 | COG2442/Duf433 | 2393421 F | 811 | Chr | AT-T | [3,16] | joint tricistronic TU2489 with slr0769 |
| slr1062 | PemK | slr1062a | hypothetical | 357724 | 7353 | Chr | AT-T | TW | TSS of TU |
| sll1092 | PIN | ssl2138 | COG5559/DUF2281 | N/K | N/K | Chr | AT-T | [16,19] | see Figure 3 |
| sll1130 | PemK | ssl2245 | hypothetical | 1049139 R | 123 | Chr | AT-T | [16,21] | potential asRNA regulation; involved in the heat shock response |
| slr1210 | PIN | slr1209 | COG2442/Duf433 | 891086 F | 2175 | Chr | AT-T | [3,16] | TSS of TU; potential asRNA regulation |
| sll1225 | PIN | ssl2420 | hypothetical | 1678733 R | 4435 | Chr | AT-T | [16] | - |
| slr1241 | MNT | slr1240 | hypothetical | 1053924 F | 1650 | Chr | AT-T | TW | TSS of TU |
| ssr1260 | HicA (YcfA) | ssr1258 | HicB (COG1598) | 3428302 F | 660 | Chr | AT-T | TW | TSS of TU |
| slr1327 | PIN3 | ssr2201 | hypothetical | 1660041 F | 28 | Chr | AT-T | [16] | - |
| ssl1377 | HicA (YcfA) | ssl1376 | HicB (COG1598) | 3427318 R | 115 | Chr | AT-T | TW | - |
| slr1383 | PIN | ssr2317 | hypothetical | 679231 F | 459 | Chr | T-AT | TW | - |
| sll1400 | PIN | ssl2733 | COG2442/Duf433 | 50262 R | 1627 | Chr | AT-T | [3,16] | TSS of TU |
| slr1421 | DUF4258 | ssr2377 | HicB (COG1598) | 3273485 F | 24 | Chr | AT-T | TW | - |
| ssl1493a | PIN | ssl1493 | PhdYeFM_antitox | 3044721 R | 66 | Chr | AT-T | TW | - |
| sll1504 | MNT | sll1505 | HEPN (DUF86) | 477658 R | 1915 | Chr | T-AT | [3,16] | TSS of TU |
| sll1543 | PIN | ssl2999 | hypothetical | 3475237 R | 53 | Chr | T-AT | TW | both components exhibit TSS |
| sll1651 | DUF497/COG2929 | sll1652 | DUF3680/COG5304 | 282821 R | 0 | Chr | T-AT | TW | leader-less |
| sll1715 | PIN | sll1714 | hypothetical | 969538 R | 244 | Chr | AT-T | [16] | - |
| ssr1766 | HicA (YcfA) | ssr1765 | HicB (COG1598) | 382338 F | 54 | Chr | AT-T | [3,16] | - |
| slr1767 | PIN | ssr2962 | COG2442/Duf433 | 544995 F | 34 | Chr | AT-T | [3,16,20] | alternative TSS (2085834 F); excludon-like arrangement |
| slr1906 | GNAT | slr1907 | hypothetical | 609442 F | 27 | Chr | T-AT | TW | - |
| sll1912 | PIN | ssl3615 | hypothetical | 776369 R | 27 | Chr | AT-T | [16] | - |
| sll1965 | DUF497/COG2929 | ssl3719 | DUF4415/COG3514 | 915052 R | 0 | Chr | T-AT | TW | leader-less |
| slr1999 | HicA (YcfA) | slr1999 | HicB (COG1598) | 1444698 | 727 | Chr | AT/T | TW | both domains in one protein |
| ssr2066 | HicA (YcfA) | ssr2067 | HicB (COG1598) | 1053924 F | 2563 | Chr | T-AT | TW | TSS of TU |
| ssl2749 | MNT | sll1411 | HEPN (DUF86) | 1619636 R | 1540 | Chr | T-AT | [3,16] | TSS of TU |
| ssr2755 | YoeB | ssr2754 | PhdYeFM_antitox | 2074673 F | 21 | Chr | AT-T | TW | - |
| N/K | N/K | ssr2784 | hypothetical (ChpI homolog) | 3474436 F | 26 | Chr | AT | TW | stand-alone |
| N/K | N/K | ssr2784a | COG2442/Duf433 | N/K | N/K | Chr | AT | TW | stand-alone; potential asRNA regulation |
| ssl2921 | HigB | ssl2920 | hypothetical | N/K | N/K | Chr | AT-T | TW | TSS of TU |
| ssl2923 | PIN | ssl2922 | VagC | N/K | N/K | Chr | AT-T | [3,16] | see Figure 3 |
| N/K | N/K | ssr3154 | HTH (COG2886) | 1211302 F | 99 | Chr | AT | TW | stand-alone; potential asRNA regulation |
| ssr3572 | YoeB | ssr3571 | PhdYeFM_antitox | 1620247 F | 29 | Chr | AT-T | TW | - |
| ssr3589 | PIN | ssr3588 | hypothetical | 1644850 F | 3314 | Chr | AT-T | TW | TSS of TU |
| sll5003 | PIN | sll5004 | COG2442/Duf433 | 1846 R | 20 | pSYSM | AT-T | [3,16] | - |
| slr5017 | MNT | slr5018 | HEPN (DUF86) | 23222 F | 33 | pSYSM | T-AT | [3,16] | - |
| ssr5020 | HigB | slr5021 | HTH-Xre | 24162 F | 19 | pSYSM | T-AT | [3,16] | - |
| N/K | N/K | ssl5025 | COG2442/Duf433 | 28079 R | 26 | pSYSM | AT | TW | stand-alone |
| sll5094 | PIN | ssl5095 | PhdYeFM_antitox | 93540 F | 20 | pSYSM | AT-T | [3,16] | - |
| slr5102 | DUF1814 | slr5101 | hypothetical | 96110 F | 14 | pSYSM | AT-T | TW | - |
| slr5116 | PIN3 | ssr5117 | HicB | 106534 F | 571 | pSYSM | T-AT | [3,16] | - |
| slr6049 | PIN | ssr6048 | hypothetical | 47161 F | 21 | pSYSX | AT-T | TW | - |
| slr6057 | COG2856/Duf955 | slr6056 | HTH-Xre | 54326 F | 17 | pSYSX | AT-T | [3,16] | potential asRNA regulation; linked to possible bacterial multiubiquitin system; see Figure 4 |
| slr6100 | RelE (DUF2136) | slr6101 | HTH-Xre | 93754 F | 66 | pSYSX | T-AT | [3,16,22] | - |
| sll7003 | PIN | ssl7004 | plasmid stability | 2074 R | 42 | pSYSA | AT-T | [3,16,23] | - |
| ssl7007 | PIN | sll7006 | HicB | 3798 R | 207 | pSYSA | T-AT | [3,16,23] | - |
| ssr7017 | cas2 | slr7016 | cas1 | N/K | N/K | pSYSA | T-AT | [33] | potential asRNA regulation |
| sll7030 | GNAT | sll7031 | COG4453/DUF1778 | 29663 R (27264 R) | 24 | pSYSA | AT-T | [3,16] | - |
| N/K | N/K | slr7032 | HEPN (DUF86) | 30317 F (27918 F) | 35 | pSYSA | AT | TW | stand-alone |
| sll7033 | PIN | sll7034 | COG2442/Duf433 | 31387 R (28988 R) | 18 | pSYSA | AT-T | [3,16,23] | - |
| ssl7039 | RelE | ssl7038 | HTH-Xre | 36479 R (34080 R) | 0 | pSYSA | T-AT | [3,16,23] | leader-less; see Figure 5 and Figure 6 |
| slr7041 | MazF | ssr7040 | MazE | 36567 F (34168 F) | 28 | pSYSA | AT-T | [3,16,23] | - |
| ssl7046 | PIN3 | N/K | N/K | 40539 R (38140 R) | 25 | pSYSA | T | TW | stand-alone |
| N/K | N/K | ssl7048 | MazE (AbrB) | N/K | N/K | pSYSA | AT | TW | stand-alone |
| ssr7072 | cas2 | slr7071 | cas1 | 64765 F (62366 F) | 57 | pSYSA | AT-T | [33] | - |
| ssr7093 | cas2 | slr7092 | cas1 | 88315 F | 312 | pSYSA | AT-T | [33] | - |
| sll8007 | PIN | ssl8008 | COG2442/Duf433 | 6626 R | 25 | pSYSG | AT-T | [3,16] | - |
| sll8011 | PIN | sll8012 | hypothetical | 9650 R | 29 | pSYSG | AT-T | [3,16] | - |
| slr8014 | PIN3 | ssr8013 | DUF2281 | 8658 F | 1055 | pSYSG | AT-T | [3,16] | see Figure 3 and Figure 7 |
| sll8027 | PIN3 | ssl8028 | MazE (AbrB) | 24116 R | 39 | pSYSG | AT-T | [3,16] | - |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopfmann, S.; Roesch, S.K.; Hess, W.R. Type II Toxin–Antitoxin Systems in the Unicellular Cyanobacterium Synechocystis sp. PCC 6803. Toxins 2016, 8, 228. https://doi.org/10.3390/toxins8070228
Kopfmann S, Roesch SK, Hess WR. Type II Toxin–Antitoxin Systems in the Unicellular Cyanobacterium Synechocystis sp. PCC 6803. Toxins. 2016; 8(7):228. https://doi.org/10.3390/toxins8070228
Chicago/Turabian StyleKopfmann, Stefan, Stefanie K. Roesch, and Wolfgang R. Hess. 2016. "Type II Toxin–Antitoxin Systems in the Unicellular Cyanobacterium Synechocystis sp. PCC 6803" Toxins 8, no. 7: 228. https://doi.org/10.3390/toxins8070228
APA StyleKopfmann, S., Roesch, S. K., & Hess, W. R. (2016). Type II Toxin–Antitoxin Systems in the Unicellular Cyanobacterium Synechocystis sp. PCC 6803. Toxins, 8(7), 228. https://doi.org/10.3390/toxins8070228