Exolysin Shapes the Virulence of Pseudomonas aeruginosa Clonal Outliers
Abstract
:1. Introduction
2. Exolysin Belongs to a Family of Two-Partner-Secretion Pore-Forming Toxins
3. Affinity of Exolysin to Membranes
4. Virulence of P. aeruginosa Strains Secreting Exolysin
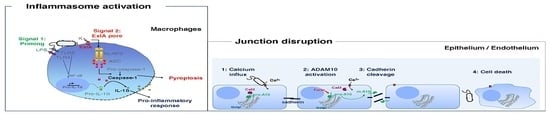
5. Caspase-1 Dependent Death of Macrophages
6. Exolysin Targets Host Cell Junctions
7. Structural Features of Exolysin
8. Conclusions and Future Directions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef]
- Cao, H.; Baldini, R.L.; Rahme, L.G. Common mechanisms for pathogens of plants and animals. Annu. Rev. Phytopathol. 2001, 39, 259–284. [Google Scholar] [CrossRef] [PubMed]
- Rahme, L.G.; Ausubel, F.M.; Cao, H.; Drenkard, E.; Goumnerov, B.C.; Lau, G.W.; Mahajan-Miklos, S.; Plotnikova, J.; Tan, M.W.; Tsongalis, J.; et al. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 2000, 97, 8815–8821. [Google Scholar] [CrossRef] [PubMed]
- Brewer, C.; Wunderink, R.G.; Jones, C.B.; Leeper, K.V. Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest 1996, 109, 1019–1029. [Google Scholar] [CrossRef]
- Kos, V.N.; Deraspe, M.; McLaughlin, R.E.; Whiteaker, J.D.; Roy, P.H.; Alm, R.A.; Corbeil, J.; Gardner, H. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob. Agents Chemother. 2015, 59, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Huber, P.; Basso, P.; Reboud, E.; Attree, I. Pseudomonas aeruginosa renews its virulence factors. Environ. Microbiol. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.H.; Tetu, S.G.; Larouche, A.; Elbourne, L.; Tremblay, S.; Ren, Q.; Dodson, R.; Harkins, D.; Shay, R.; Watkins, K.; et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE 2010, 5, e8842. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [PubMed]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.X.; Saucier, M.; Deziel, E.; et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Pseudomonas Database. Available online: www.pseudomonas.com (accessed on 9 November 2017).
- Wiehlmann, L.; Cramer, N.; Tummler, B. Habitat-associated skew of clone abundance in the Pseudomonas aeruginosa population. Environ. Microbiol. Rep. 2015, 7, 955–960. [Google Scholar] [CrossRef] [PubMed]
- Thrane, S.W.; Taylor, V.L.; Freschi, L.; Kukavica-Ibrulj, I.; Boyle, B.; Laroche, J.; Pirnay, J.P.; Levesque, R.C.; Lam, J.S.; Jelsbak, L. The Widespread Multidrug-Resistant Serotype O12 Pseudomonas aeruginosa Clone Emerged through Concomitant Horizontal Transfer of Serotype Antigen and Antibiotic Resistance Gene Clusters. mBio 2015, 6, e01396-15. [Google Scholar] [CrossRef] [PubMed]
- Hilker, R.; Munder, A.; Klockgether, J.; Losada, P.M.; Chouvarine, P.; Cramer, N.; Davenport, C.F.; Dethlefsen, S.; Fischer, S.; Peng, H.; et al. Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ. Microbiol. 2015, 17, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R. The type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat. Rev. Microbiol. 2009, 7, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Filloux, A. Protein Secretion Systems in Pseudomonas aeruginosa: An Essay on Diversity, Evolution, and Function. Front. Microbiol. 2011, 2, 155. [Google Scholar] [CrossRef] [PubMed]
- Berthelot, P.; Attree, I.; Plesiat, P.; Chabert, J.; de Bentzmann, S.; Pozzetto, B.; Grattard, F.; Groupe d’Etudes des Septicémies à Pseudomonas aeruginosa. Genotypic and phenotypic analysis of type III secretion system in a cohort of Pseudomonas aeruginosa bacteremia isolates: Evidence for a possible association between O serotypes and exo genes. J. Infect. Dis. 2003, 188, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Shaver, C.M.; Hauser, A.R. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 2004, 72, 6969–6977. [Google Scholar] [CrossRef] [PubMed]
- Reboud, E.; Elsen, S.; Bouillot, S.; Golovkine, G.; Basso, P.; Jeannot, K.; Attree, I.; Huber, P. Phenotype and toxicity of the recently discovered exlA-positive Pseudomonas aeruginosa strains collected worldwide. Environ. Microbiol. 2016, 18, 3425–3439. [Google Scholar] [CrossRef] [PubMed]
- Elsen, S.; Huber, P.; Bouillot, S.; Coute, Y.; Fournier, P.; Dubois, Y.; Timsit, J.F.; Maurin, M.; Attree, I. A type III secretion negative clinical strain of Pseudomonas aeruginosa employs a two-partner secreted exolysin to induce hemorrhagic pneumonia. Cell Host Microbe 2014, 15, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Basso, P.; Wallet, P.; Elsen, S.; Soleilhac, E.; Henry, T.; Faudry, F.; Attrée, I. Multiple Pseudomonas species secrete Exolysin-like toxins and provoke Caspase-1-dependent macrophage death. Environ. Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Basso, P.; Ragno, M.; Elsen, S.; Reboud, E.; Golovkine, G.; Bouillot, S.; Huber, P.; Lory, S.; Faudry, E.; Attree, I. Pseudomonas aeruginosa Pore-Forming Exolysin and Type IV Pili Cooperate To Induce Host Cell Lysis. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Dubuisson, F.; Guerin, J.; Baelen, S.; Clantin, B. Two-partner secretion: as simple as it sounds? Res. Microbiol. 2013, 164, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Dubuisson, F.; Locht, C.; Antoine, R. Two-partner secretion in Gram-negative bacteria: A thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 2001, 40, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Dubuisson, F.; Villeret, V.; Clantin, B.; Delattre, A.S.; Saint, N. First structural insights into the TpsB/Omp85 superfamily. Biol. Chem. 2009, 390, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Ondraczek, R.; Hobbie, S.; Braun, V. In vitro activation of the Serratia marcescens hemolysin through modification and complementation. J. Bacteriol. 1992, 174, 5086–5094. [Google Scholar] [CrossRef] [PubMed]
- Braun, V.; Neuss, B.; Ruan, Y.; Schiebel, E.; Schoffler, H.; Jander, G. Identification of the Serratia marcescens hemolysin determinant by cloning into Escherichia coli. J. Bacteriol. 1987, 169, 2113–2120. [Google Scholar] [CrossRef] [PubMed]
- Schiebel, E.; Schwarz, H.; Braun, V. Subcellular location and unique secretion of the hemolysin of Serratia marcescens. J. Biol. Chem. 1989, 264, 16311–16320. [Google Scholar] [PubMed]
- Weaver, T.M.; Smith, J.A.; Hocking, J.M.; Bailey, L.J.; Wawrzyn, G.T.; Howard, D.R.; Sikkink, L.A.; Ramirez-Alvarado, M.; Thompson, J.R. Structural and functional studies of truncated hemolysin A from Proteus mirabilis. J. Biol. Chem. 2009, 284, 22297–22309. [Google Scholar] [CrossRef] [PubMed]
- Schiebel, E.; Braun, V. Integration of the Serratia marcescens haemolysin into human erythrocyte membranes. Mol. Microbiol. 1989, 3, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Schonherr, R.; Hilger, M.; Broer, S.; Benz, R.; Braun, V. Interaction of Serratia marcescens hemolysin (ShlA) with artificial and erythrocyte membranes. Demonstration of the formation of aqueous multistate channels. Eur. J. Biochem. 1994, 223, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Reboud, E.; Bouillot, S.; Patot, S.; Beganton, B.; Attree, I.; Huber, P. Pseudomonas aeruginosa ExIA and Serratia marcescens ShIA trigger cadherin cleavage by promoting calcium influx and ADAM10 activation. PLoS Pathog. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Dal Peraro, M.; van der Goot, F.G. Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016, 14, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Faudry, E.; Vernier, G.; Neumann, E.; Forge, V.; Attree, I. Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa. Biochemistry 2006, 45, 8117–8123. [Google Scholar] [CrossRef] [PubMed]
- Hertle, R. Serratia marcescens hemolysin (ShIA) binds artificial membranes and forms pores in a receptor-independent manner. J. Membr. Biol. 2002, 189, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bouillot, S.; Munro, P.; Gallet, B.; Reboud, E.; Cretin, F.; Golovkine, G.; Schoehn, G.; Attree, I.; Lemichez, E.; Huber, P. Pseudomonas aeruginosa Exolysin promotes bacterial growth in lungs, alveolar damage and bacterial dissemination. Sci. Rep. 2017, 7, 2120. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Juarbe, N.; Mares, C.A.; Hinojosa, C.A.; Medina, J.L.; Cantwell, A.; Dube, P.H.; Orihuela, C.J.; Bergman, M.A. Requirement for Serratia marcescens cytolysin in a murine model of hemorrhagic pneumonia. Infect. Immun. 2015, 83, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Suarez Mdel, M.; Florez, N.; Astudillo, A.; Vazquez, F.; Villaverde, R.; Fabrizio, K.; Pirofski, L.A.; Mendez, F.J. The role of pneumolysin in mediating lung damage in a lethal pneumococcal pneumonia murine model. Respir. Res. 2007, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Diep, B.A.; Chan, L.; Tattevin, P.; Kajikawa, O.; Martin, T.R.; Basuino, L.; Mai, T.T.; Marbach, H.; Braughton, K.R.; Whitney, A.R.; et al. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc. Natl. Acad. Sci. USA 2010, 107, 5587–5592. [Google Scholar] [CrossRef] [PubMed]
- Greaney, A.J.; Leppla, S.H.; Moayeri, M. Bacterial exotoxins and the inflammasome. Front. Immunol. 2015, 6, 570. [Google Scholar] [CrossRef] [PubMed]
- Ebsen, H.; Lettau, M.; Kabelitz, D.; Janssen, O. Identification of SH3 Domain Proteins Interacting with the Cytoplasmic Tail of the A Disintegrin and Metalloprotease 10 (ADAM10). PLoS ONE 2014, 9, e102899. [Google Scholar] [CrossRef] [PubMed]
- Seals, D.F.; Courtneidge, S.A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003, 17, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.; Le Gall, S.; Schulte, M.; Yamaguchi, T.; Reiss, K.; Murphy, G.; Toyama, Y.; Hartmann, D.; Saftig, P.; Blobel, C.P. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol. Biol. Cell 2007, 18, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Nagano, O.; Murakami, D.; Hartmann, D.; de Strooper, B.; Saftig, P.; Iwatsubo, T.; Nakajima, M.; Shinohara, M.; Saya, H. Cell-matrix interaction via CD44 is independently regulated by different metal loproteinases activated in response to extracellular Ca2+ influx and PKC activation. J. Cell Biol. 2004, 165, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Inoshima, I.; Inoshima, N.; Wilke, G.A.; Powers, M.E.; Frank, K.M.; Wang, Y.; Wardenburg, J.B. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 2011, 17, U1310–U1314. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.E.; Kim, H.K.; Wang, Y.; Wardenburg, J.B. ADAM10 Mediates Vascular Injury Induced by Staphylococcus aureus alpha-Hemolysin. J. Infect. Dis. 2012, 206, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Poole, K.; Schiebel, E.; Braun, V. Molecular characterization of the hemolysin determinant of Serratia marcescens. J. Bacteriol. 1988, 170, 3177–3188. [Google Scholar] [CrossRef] [PubMed]
- Guérin, J.; Bigot, S.; Schneider, R.; Buchanan, S.K.; Jacob-Dubuisson, F. Two-Partner Secretion: Combining Efficiency and Simplicity in the Secretion of Large Proteins for Bacteria-Host and Bacteria-Bacteria Interactions. Front. Cell. Infect. Microbiol. 2017, 7, 148. [Google Scholar] [CrossRef] [PubMed]
- Zambolin, S.; Clantin, B.; Chami, M.; Hoos, S.; Haouz, A.; Villeret, V.; Delepelaire, P. Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat. Commun. 2016, 7, 11590. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Relman, D.A.; Nishikawa, A. Invasion of human respiratory epithelial cells by Bordetella pertussis: Possible role for a filamentous hemagglutinin Arg-Gly-Asp sequence and alpha 5 beta 1 integrin. Microb. Pathog. 2001, 30, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Julio, S.M.; Inatsuka, C.S.; Mazar, J.; Dieterich, C.; Relman, D.A.; Cotter, P.A. Natural-host animal models indicate functional interchangeability between the filamentous haemagglutinins of Bordetella pertussis and Bordetella bronchiseptica and reveal a role for the mature C-terminal domain, but not the RGD motif, during infection. Mol. Microbiol. 2009, 71, 1574–1590. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Clantin, B.; Hodak, H.; Willery, E.; Locht, C.; Jacob-Dubuisson, F.; Villeret, V. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 6194–6199. [Google Scholar] [CrossRef] [PubMed]
- Yeo, H.J.; Yokoyama, T.; Walkiewicz, K.; Kim, Y.; Grass, S.; Geme, J.W., 3rd. The structure of the Haemophilus influenzae HMW1 pro-piece reveals a structural domain essential for bacterial two-partner secretion. J. Biol. Chem. 2007, 282, 31076–31084. [Google Scholar] [CrossRef] [PubMed]
- Kajava, A.V.; Steven, A.C. The turn of the screw: variations of the abundant beta-solenoid motif in passenger domains of Type V secretory proteins. J. Struct. Biol. 2006, 155, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.; Hertle, R.; Braun, V. Activation of Serratia marcescens hemolysin through a conformational change. Infect. Immun. 2004, 72, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Los, F.C.O.; Randis, T.M.; Aroian, R.V.; Ratner, A.J. Role of Pore-Forming Toxins in Bacterial Infectious Diseases. Microbiol. Mol. Biol. Rev. 2013, 77, 173–207. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, S.K.B.; O’Riordan, M.X.D. More Than a Pore: The Cellular Response to Cholesterol-Dependent Cytolysins. Toxins 2013, 5, 618–636. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Keyel, M.; Shi, G.L.; Bhattacharjee, P.; Roth, R.; Heuser, J.E.; Keyel, P.A. Intrinsic repair protects cells from pore-forming toxins by microvesicle shedding. Cell Death Differ. 2017, 24, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Von Hoven, G.; Rivas, A.J.; Neukirch, C.; Meyenburg, M.; Qin, Q.; Parekh, S.; Hellmann, N.; Husmann, M. Repair of a Bacterial Small beta-Barrel Toxin Pore Depends on Channel Width. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reboud, E.; Basso, P.; Maillard, A.P.; Huber, P.; Attrée, I. Exolysin Shapes the Virulence of Pseudomonas aeruginosa Clonal Outliers. Toxins 2017, 9, 364. https://doi.org/10.3390/toxins9110364
Reboud E, Basso P, Maillard AP, Huber P, Attrée I. Exolysin Shapes the Virulence of Pseudomonas aeruginosa Clonal Outliers. Toxins. 2017; 9(11):364. https://doi.org/10.3390/toxins9110364
Chicago/Turabian StyleReboud, Emeline, Pauline Basso, Antoine P. Maillard, Philippe Huber, and Ina Attrée. 2017. "Exolysin Shapes the Virulence of Pseudomonas aeruginosa Clonal Outliers" Toxins 9, no. 11: 364. https://doi.org/10.3390/toxins9110364
APA StyleReboud, E., Basso, P., Maillard, A. P., Huber, P., & Attrée, I. (2017). Exolysin Shapes the Virulence of Pseudomonas aeruginosa Clonal Outliers. Toxins, 9(11), 364. https://doi.org/10.3390/toxins9110364