Organs-on-a-Chip Module: A Review from the Development and Applications Perspective
Abstract
:1. Introduction
2. Microfluidics Techniques
Advantages and Challenges: Traditional vs. Microfluidic Cell Culture Approach
3. Three-Dimensional (3-D) Printed Microfluidics
4. Potential Materials and Fabrication Techniques
5. Electrokinetic Phenomena: Theory and Microfluidic Applications
6. Microfluidic: Lab-on-a-Chip
7. Microfluidic: Organ-on-a-Chip
7.1. Lung-on-a-Chip
7.2. Liver-on-a-Chip
7.3. Kidney-on-a-Chip
7.4. Gut-on-a-Chip
7.5. Skin-on-a-Chip
7.6. Brain-on-a-Chip
7.7. Heart-on-a-Chip
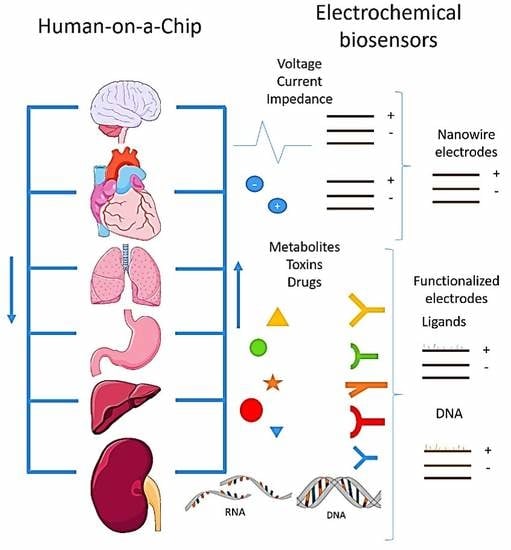
8. Human-on-a-Chip
9. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manz, A.; Graber, N.; Widmer, H.Á. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Verpoorte, E.; De Rooij, N.F. Microfluidics meets MEMS. Proc. IEEE 2003, 91, 930–953. [Google Scholar] [CrossRef]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Bravard, J.P.; Petit, F. Geomorphology of Streams and Rivers. In Encyclopedia of Inland Waters; Elsevier: Amsterdam, The Netherlands, 2009; pp. 387–395. [Google Scholar]
- Mohammed, M.I.; Zainal Alam, M.N.H.; Kouzani, A.; Gibson, I. Fabrication of Microfluidic Devices: Improvement of Surface Quality of CO2 Laser Machined Poly(Methylmethacrylate) Polymer. J. Micromech. Microeng. 2017, 27, 015021. [Google Scholar] [CrossRef]
- Hong, T.-F.; Ju, W.-J.; Wu, M.-C.; Tai, C.-H.; Tsai, C.-H.; Fu, L.-M. Rapid Prototyping of PMMA Microfluidic Chips Utilizing a CO2 Laser. Microfluid. Nanofluid. 2010, 9, 1125–1133. [Google Scholar] [CrossRef]
- Owens, C.E.; Hart, A.J. High-Precision Modular Microfluidics by Micromilling of Interlocking Injection-Molded Blocks. Lab Chip 2018, 18, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic Lab-on-a-Chip Platforms: Requirements, Characteristics and Applications. Chem. Soc. Rev. 2010, 39, 1153. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Iqbal, H.M.N.; Akram, Z. Microfluidics Engineering: Recent Trends, Valorization, and Applications. Arab. J. Sci. Eng. 2018, 43, 23–32. [Google Scholar] [CrossRef]
- Whitesides, G.M.; Ostuni, E.; Takayama, S.; Jiang, X.; Ingber, D.E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001, 3, 335–373. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.K.; Whitesides, G.M. Microfluidic devices fabricated in poly (dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Paguirigan, A.L.; Beebe, D.J. Microfluidics meet cell biology: Bridging the gap by validation and application of microscale techniques for cell biological assays. Bioessays 2008, 30, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.B.; Ng, S.H.; Li, K.H.H.; Yoon, Y.J. 3D printed microfluidics for biological applications. Lab Chip 2015, 15, 3627–3637. [Google Scholar] [CrossRef] [PubMed]
- Gross, B.C.; Erkal, J.L.; Lockwood, S.Y.; Chen, C.; Spence, D.M. Evaluation of 3D printing and its potential impact on biotechnology and the chemical sciences. Anal. Chem. 2014, 86, 3240–3253. [Google Scholar] [CrossRef] [PubMed]
- Serex, L.; Bertsch, A.; Renaud, P. Microfluidics: A New Layer of Control for Extrusion-Based 3D Printing. Micromachines 2018, 9, 86. [Google Scholar] [CrossRef]
- Tsao, C.-W. Polymer Microfluidics: Simple, Low-Cost Fabrication Process Bridging Academic Lab Research to Commercialized Production. Micromachines 2016, 7, 225. [Google Scholar] [CrossRef]
- Ahmed, I.; Akram, Z.; Bule, M.H.; Iqbal, H.M.N. Advancements and Potential Applications of Microfluidic Approaches—A Review. Chemosensors 2018, 6, 46. [Google Scholar] [CrossRef]
- Edington, C.D.; Chen, W.L.K.; Geishecker, E.; Kassis, T.; Soenksen, L.R.; Bhushan, B.M.; Valdez, J. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Sci. Rep. 2018, 8, 4530. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Wu, L.-Q.; Ghodssi, R.; Rubloff, G.W.; Payne, G.F.; Bentley, W.E. Signal-Directed Sequential Assembly of Biomolecules on Patterned Surfaces. Langmuir 2005, 21, 2104–2107. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Wu, L.-Q.; Bentley, W.E.; Ghodssi, R.; Rubloff, G.W.; Culver, J.N.; Payne, G.F. Biofabrication with Chitosan. Biomacromolecules 2005, 6, 2881–2894. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, C.J.; Cyr, K.M.; Matsumoto, A.; Langer, R.; Borenstein, J.T.; Kaplan, D.L. Silk Fibroin Microfluidic Devices. Adv. Mater. 2007, 19, 2847–2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Rigueiro, J.; Viney, C.; Llorca, J.; Elices, M. Mechanical Properties of Single-Brin Silkworm Silk. J. Appl. Polym. Sci. 2000, 75, 1270–1277. [Google Scholar] [CrossRef]
- Ling, Y.; Rubin, J.; Deng, Y.; Huang, C.; Demirci, U.; Karp, J.M.; Khademhosseini, A. A Cell-Laden Microfluidic Hydrogel. Lab Chip 2007, 7, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Masuda, K.; Sah, R.L.; Hejna, M.J.; Thonar, E.J.-M.A. A Novel Two-Step Method for the Formation of Tissue-Engineered Cartilage by Mature Bovine Chondrocytes: The Alginate-Recovered-Chondrocyte (ARC) Method. J. Orthop. Res. 2003, 21, 139–148. [Google Scholar] [CrossRef]
- Rahfoth, B.; Weisser, J.; Sternkopf, F.; Aigner, T.; von der Mark, K.; Bräuer, R. Transplantation of Allograft Chondrocytes Embedded in Agarose Gel into Cartilage Defects of Rabbits. Osteoarthr. Cartil. 1998, 6, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Grover, W.H.; Von Muhlen, M.G.; Manalis, S.R. Teflon Films for Chemically-Inert Microfluidic Valves and Pumps. Lab Chip 2008, 8, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Jiang, H. Autonomous Microfluidics with Stimuli-Responsive Hydrogels. Soft Matter 2007, 3, 1223. [Google Scholar] [CrossRef]
- Rogers, C.I.; Qaderi, K.; Woolley, A.T.; Nordin, G.P. 3D Printed Microfluidic Devices with Integrated Valves. Biomicrofluidics 2015, 9, 016501. [Google Scholar] [CrossRef] [PubMed]
- Shallan, A.I.; Smejkal, P.; Corban, M.; Guijt, R.M.; Breadmore, M.C. Cost-Effective Three-Dimensional Printing of Visibly Transparent Microchips within Minutes. Anal. Chem. 2014, 86, 3124–3130. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Shin, J.H.; Han, K.-H. An On-Chip RT-PCR Microfluidic Device, That Integrates MRNA Extraction, CDNA Synthesis, and Gene Amplification. RSC Adv. 2014, 4, 9160. [Google Scholar] [CrossRef]
- Lee, H.; Han, N.; Choi, I.-H.; Han, K.-H. A High-Speed, High-Performance on-Chip Integrated Reverse Transcription (RT)-Microchip. Biomed. Microdevices 2013, 15, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Macdonald, N.P.; Cooper, J.M.; Wlodkowic, D. Additive Manufacturing of Lab-on-a-Chip Devices: Promises and Challenges. In Micro/Nano Materials, Devices, and Systems; Friend, J., Tan, H.H., Eds.; International Society for Optics and Photonics: Melbourne, Australia, 2013; Volume 8923, p. 892344. [Google Scholar]
- Sugioka, K.; Hanada, Y.; Midorikawa, K. 3D microstructuring of glass by femtosecond laser direct writing and application to biophotonic microchips. Prog. Electromagn. Res. Lett. 2008, 1, 181–188. [Google Scholar] [CrossRef]
- Hanada, Y.; Sugioka, K.; Shihira-Ishikawa, I.; Kawano, H.; Miyawaki, A.; Midorikawa, K. 3D Microfluidic Chips with Integrated Functional Microelements Fabricated by a Femtosecond Laser for Studying the Gliding Mechanism of Cyanobacteria. Lab Chip 2011, 11, 2109–2115. [Google Scholar] [CrossRef] [PubMed]
- Chudobova, D.; Cihalova, K.; Skalickova, S.; Zitka, J.; Rodrigo, M.A.M.; Milosavljevic, V.; Hynek, D.; Kopel, P.; Vesely, R.; Adam, V.; et al. 3D-Printed Chip for Detection of Methicillin-Resistant Staphylococcus Aureus Labeled with Gold Nanoparticles. Electrophoresis 2015, 36, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Krejcova, L.; Nejdl, L.; Rodrigo, M.A.M.; Zurek, M.; Matousek, M.; Hynek, D.; Zitka, O.; Kopel, P.; Adam, V.; Kizek, R. 3D Printed Chip for Electrochemical Detection of Influenza Virus Labeled with CdS Quantum Dots. Biosens. Bioelectron. 2014, 54, 421–427. [Google Scholar] [CrossRef] [PubMed]
- King, P.H.; Jones, G.; Morgan, H.; de Planque, M.R.R.; Zauner, K.-P. Interdroplet Bilayer Arrays in Millifluidic Droplet Traps from 3D-Printed Moulds. Lab Chip 2014, 14, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.; Gärtner, C. Polymer Microfabrication Technologies for Microfluidic Systems. Anal. Bioanal. Chem. 2008, 390, 89–111. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.S.; Stevens, K.R.; Yang, M.T.; Baker, B.M.; Nguyen, D.-H.T.; Cohen, D.M.; Toro, E.; Chen, A.A.; Galie, P.A.; Yu, X.; et al. Rapid Casting of Patterned Vascular Networks for Perfusable Engineered Three-Dimensional Tissues. Nat. Mater. 2012, 11, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Bertassoni, L.E.; Cecconi, M.; Manoharan, V.; Nikkhah, M.; Hjortnaes, J.; Cristino, A.L.; Barabaschi, G.; Demarchi, D.; Dokmeci, M.R.; Yang, Y.; et al. Hydrogel Bioprinted Microchannel Networks for Vascularization of Tissue Engineering Constructs. Lab Chip 2014, 14, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.S.; Ng, L.; Yen, G.S.; Lorenz, R.M.; Schiro, P.G.; Edgar, J.S.; Zhao, Y.; Lim, D.S.W.; Allen, P.B.; Jeffries, G.D.M.; et al. A New USP Class VI-Compliant Substrate for Manufacturing Disposable Microfluidic Devices. Lab Chip 2009, 9, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.I.; Oxborrow, J.B.; Anderson, R.R.; Tsai, L.-F.; Nordin, G.P.; Woolley, A.T. Microfluidic Valves Made from Polymerized Polyethylene Glycol Diacrylate. Sens. Actuators B Chem. 2014, 191, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Plegue, T.J.; Kovach, K.M.; Thompson, A.J.; Potkay, J.A. Stability of Polyethylene Glycol and Zwitterionic Surface Modifications in PDMS Microfluidic Flow Chambers. Langmuir 2018, 34, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Cherpinski, A.; Torres-Giner, S.; Vartiainen, J.; Peresin, M.S.; Lahtinen, P.; Lagaron, J.M. Improving the Water Resistance of Nanocellulose-Based Films with Polyhydroxyalkanoates Processed by the Electrospinning Coating Technique. Cellulose 2018, 25, 1291–1307. [Google Scholar] [CrossRef]
- Chen, M.B.; Srigunapalan, S.; Wheeler, A.R.; Simmons, C.A. A 3D Microfluidic Platform Incorporating Methacrylated Gelatin Hydrogels to Study Physiological Cardiovascular Cell–cell Interactions. Lab Chip 2013, 13, 2591–2598. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.; Kim, S.H.; Lee, D.; Kim, B.; Kim, T.H.; Jung, Y.; Choi, N.; Sung, J.H. Fabrication of Micrometer-Scale Porous Gelatin Scaffolds for 3D Cell Culture. J. Ind. Eng. Chem. 2017, 50, 183–189. [Google Scholar] [CrossRef]
- Zamboni, F.; Keays, M.; Hayes, S.; Albadarin, A.B.; Walker, G.M.; Kiely, P.A.; Collins, M.N. Enhanced Cell Viability in Hyaluronic Acid Coated Poly(Lactic-Co-Glycolic Acid) Porous Scaffolds within Microfluidic Channels. Int. J. Pharm. 2017, 532, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Mogosanu, D.-E.; Verplancke, R.; Dubruel, P.; Vanfleteren, J. Fabrication of 3-Dimensional Biodegradable Microfluidic Environments for Tissue Engineering Applications. Mater. Des. 2016, 89, 1315–1324. [Google Scholar] [CrossRef]
- Miri, A.K.; Nieto, D.; Iglesias, L.; Goodarzi Hosseinabadi, H.; Maharjan, S.; Ruiz-Esparza, G.U.; Shin, S.R. Microfluidics-Enabled Multimaterial Maskless Stereolithographic Bioprinting. Adv. Mater. 2018, 30, 1800242. [Google Scholar] [CrossRef] [PubMed]
- Bertana, V.; Potrich, C.; Scordo, G.; Scaltrito, L.; Ferrero, S.; Lamberti, A.; Marasso, S.L. 3D-printed microfluidics on thin poly (methyl methacrylate) substrates for genetic applications. J. Vac. Sci. Technol. B Nanotechnol. Microelectron. Mater. Process. Meas. Phenom. 2018, 36, 01A106. [Google Scholar] [CrossRef]
- Roy, E.; Geissler, M.; Galas, J.C.; Veres, T. Prototyping of microfluidic systems using a commercial thermoplastic elastomer. Microfluid. Nanofluid. 2011, 11, 235–244. [Google Scholar] [CrossRef]
- Borysiak, M.D.; Yuferova, E.; Posner, J.D. Simple, low-cost styrene-ethylene/butylene-styrene microdevices for electrokinetic applications. Anal. Chem. 2013, 85, 11700–11704. [Google Scholar] [CrossRef] [PubMed]
- Domansky, K.; Sliz, J.D.; Wen, N.; Hinojosa, C.; Thompson, G.; Fraser, J.P.; Ingber, D.E. SEBS elastomers for fabrication of microfluidic devices with reduced drug absorption by injection molding and extrusion. Microfluid. Nanofluid. 2017, 21, 107. [Google Scholar] [CrossRef]
- Todd, R.H.; Allen, D.K.; Alting, L. Manufacturing Processes Reference Guide; Industrial Press Inc.: New York, NY, USA, 1994. [Google Scholar]
- Becker, H.; Locascio, L.E. Polymer microfluidic devices. Talanta 2002, 56, 267–287. [Google Scholar] [CrossRef]
- Kellogg, G.J.; Arnold, T.E.; Carvalho, B.L.; Duffy, D.C.; Sheppard, N.F. Centrifugal microfluidics: Applications. In Micro Total Analysis Systems; Springer: Dordrecht, The Netherland, 2000; pp. 239–242. [Google Scholar]
- Anderson, J.L. Colloid Transport by Interfacial Forces. Annu. Rev. Fluid Mech. 1989, 21, 61–99. [Google Scholar] [CrossRef]
- Garza-García, L.D.; Pérez-González, V.H.; Pérez-Sánchez, O.A.; Lapizco-Encinas, B.H. Electrokinetic Mobilities Characterization and Rapid Detection of Microorganisms in Glass Microchannels. Chem. Eng. Technol. 2011, 34, 371–378. [Google Scholar] [CrossRef]
- Polniak, D.V.; Goodrich, E.; Hill, N.; Lapizco-Encinas, B.H. Separating Large Microscale Particles by Exploiting Charge Differences with Dielectrophoresis. J. Chromatogr. A 2018, 1545, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Romero-Creel, M.; Goodrich, E.; Polniak, D.; Lapizco-Encinas, B. Assessment of Sub-Micron Particles by Exploiting Charge Differences with Dielectrophoresis. Micromachines 2017, 8, 239. [Google Scholar] [CrossRef]
- Squires, T.M.; Bazant, M.Z. Induced-Charge Electro-Osmosis. J. Fluid Mech. 2004, 509, 217–252. [Google Scholar] [CrossRef]
- Saucedo-Espinosa, M.A.; Rauch, M.M.; Lalonde, A.; Lapizco-Encinas, B.H. Polarization Behavior of Polystyrene Particles under Direct Current and Low-Frequency (<1 KHz) Electric Fields in Dielectrophoretic Systems. Electrophoresis 2016, 37, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Gangwal, S.; Cayre, O.J.; Bazant, M.Z.; Velev, O.D. Induced-Charge Electrophoresis of Metallodielectric Particles. Phys. Rev. Lett. 2008, 100, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lang, Q.; Wu, Y.; Ren, Y.; Tao, Y.; Lei, L.; Jiang, H. AC Electrothermal Circulatory Pumping Chip for Cell Culture. ACS Appl. Mater. Interfaces 2015, 7, 26792–26801. [Google Scholar] [CrossRef] [PubMed]
- Rashidi, S.; Bafekr, H.; Valipour, M.S.; Esfahani, J.A. A Review on the Application, Simulation, and Experiment of the Electrokinetic Mixers. Chem. Eng. Process. Process Intensif. 2018, 126, 108–122. [Google Scholar] [CrossRef]
- Li, M.; Li, D. Microvalve Using Electrokinetic Motion of Electrically Induced Janus Droplet. Anal. Chim. Acta 2018, 1021, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Shaegh, S.A.M.; Pourmand, A.; Nabavinia, M.; Avci, H.; Tamayol, A.; Mostafalu, P.; Zhang, Y.S. Rapid prototyping of whole-thermoplastic microfluidics with built-in microvalves using laser ablation and thermal fusion bonding. Sens. Actuators B Chem. 2018, 255, 100–109. [Google Scholar] [CrossRef]
- Pourmand, A.; Shaegh, S.A.M.; Ghavifekr, H.B.; Aghdam, E.N.; Dokmeci, M.R.; Khademhosseini, A.; Zhang, Y.S. Fabrication of whole-thermoplastic normally closed microvalve, micro check valve, and micropump. Sens. Actuators B Chem. 2018, 262, 625–636. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, S.; Zhang, L.; Yao, Z.; Chen, X.; Zheng, Y.; Liu, Y. Applications and Theory of Electrokinetic Enrichment in Micro-Nanofluidic Chips. Biomed. Microdevices 2017, 19, 19. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, T.; Lamanda, A.C.; Sin, M.L.Y.; Gau, V.; Liao, J.C.; Wong, P.K. AC Electrokinetics of Physiological Fluids for Biomedical Applications. J. Lab. Autom. 2015, 20, 611–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Xing, D.; Li, Y. Micropumps, microvalves, and micromixers within PCR microfluidic chips: Advances and trends. Biotechnol. Adv. 2007, 25, 483–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, C.; Wang, S.; Liu, S. Electroosmotic pumps and their applications in microfluidic systems. Microfluid. Nanofluid. 2009, 6, 145–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iverson, B.D.; Garimella, S.V. Recent advances in microscale pumping technologies: A review and evaluation. Microfluid. Nanofluid. 2008, 5, 145–174. [Google Scholar] [CrossRef]
- Carminati, M.; Ferrari, G.; Vahey, M.D.; Voldman, J.; Sampietro, M. Miniaturized Impedance Flow Cytometer: Design Rules and Integrated Readout. IEEE Trans. Biomed. Circuits Syst. 2017, 11, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Alazzam, A.; Khashan, S.; Abutayeh, M. Lab-on-Chip for Liquid Biopsy (LoC-LB) Based on Dielectrophoresis. Talanta 2017, 164, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, J.P. The relevance and potential roles of microphysiological systems in biology and medicine. Exp. Biol. Med. 2014, 239, 1061–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Stucki, A.O.; Stucki, J.D.; Hall, S.R.R.; Felder, M.; Mermoud, Y.; Schmid, R.A.; Geiser, T.; Guenat, O.T. A Lung-on-a-Chip Array with an Integrated Bio-Inspired Respiration Mechanism. Lab Chip 2015, 15, 1302–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humayun, M.; Chow, C.-W.; Young, E.W.K. Microfluidic Lung Airway-on-a-Chip with Arrayable Suspended Gels for Studying Epithelial and Smooth Muscle Cell Interactions. Lab Chip 2018, 18, 1298–1309. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Selva Kumar, N.D.; Choudhury, D.; Foo, L.C.; Ng, S.H. Microfluidic Platforms for Modeling Biological Barriers in the Circulatory System. Drug Discov. Today 2018, 23, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Barrile, R.; van der Meer, A.; Mammoto, A.; Mammoto, T.; De Ceunynck, K.; Aisiku, O.; Otieno, M.; Louden, C.; Hamilton, G.; et al. Primary Human Lung Alveolus-on-a-Chip Model of Intravascular Thrombosis for Assessment of Therapeutics. Clin. Pharmacol. Ther. 2018, 103, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-A.; No, D.Y.; Kang, E.; Ju, J.; Kim, D.-S.; Lee, S.-H. Spheroid-Based Three-Dimensional Liver-on-a-Chip to Investigate Hepatocyte–hepatic Stellate Cell Interactions and Flow Effects. Lab Chip 2013, 13, 3529–3537. [Google Scholar] [CrossRef] [PubMed]
- Yoon No, D.; Lee, K.-H.; Lee, J.; Lee, S.-H. 3D Liver Models on a Microplatform: Well-Defined Culture, Engineering of Liver Tissue and Liver-on-a-Chip. Lab Chip 2015, 15, 3822–3837. [Google Scholar] [CrossRef] [PubMed]
- Bavli, D.; Prill, S.; Ezra, E.; Levy, G.; Cohen, M.; Vinken, M.; Vanfleteren, J.; Jaeger, M.; Nahmias, Y. Real-Time Monitoring of Metabolic Function in Liver-on-Chip Microdevices Tracks the Dynamics of Mitochondrial Dysfunction. Proc. Natl. Acad. Sci. USA 2016, 113, E2231–E2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delalat, B.; Cozzi, C.; Rasi Ghaemi, S.; Polito, G.; Kriel, F.H.; Michl, T.D.; Harding, F.J.; Priest, C.; Barillaro, G.; Voelcker, N.H. Microengineered Bioartificial Liver Chip for Drug Toxicity Screening. Adv. Funct. Mater. 2018, 28, 1801825. [Google Scholar] [CrossRef]
- Musah, S.; Mammoto, A.; Ferrante, T.C.; Jeanty, S.S.F.; Hirano-Kobayashi, M.; Mammoto, T.; Roberts, K.; Chung, S.; Novak, R.; Ingram, M.; et al. Mature Induced-Pluripotent-Stem-Cell-Derived Human Podocytes Reconstitute Kidney Glomerular-Capillary-Wall Function on a Chip. Nat. Biomed. Eng. 2017, 1, 0069. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.-Y.; Ingber, D.E. Human Kidney Proximal Tubule-on-a-Chip for Drug Transport and Nephrotoxicity Assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Wilmer, M.J.; Ng, C.P.; Lanz, H.L.; Vulto, P.; Suter-Dick, L.; Masereeuw, R. Kidney-on-a-chip technology for drug-induced nephrotoxicity screening. Trends Biotechnol. 2016, 34, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human Gut-on-a-Chip Inhabited by Microbial Flora That Experiences Intestinal Peristalsis-like Motions and Flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip Microenvironment Induces Human Intestinal Cells to Undergo Villus Differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of Microbiome and Mechanical Deformation to Intestinal Bacterial Overgrowth and Inflammation in a Human Gut-on-a-Chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Romaine, P.L.; Newman, V.L. Biologics as Countermeasures for Acute Radiation Syndrome: Where Are We Now? Expert Opin. Biol. Ther. 2015, 15, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Prantil-Baun, R.; Jiang, A.; Potla, R.; Mammoto, T.; Weaver, J.C.; Ferrante, T.C.; Kim, H.J.; Cabral, J.M.S.; Levy, O.; et al. Modeling Radiation Injury-Induced Cell Death and Countermeasure Drug Responses in a Human Gut-on-a-Chip. Cell Death Dis. 2018, 9, 223. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, M.E.; Khetsuriani, N.; Fowlkes, A.L.; Zheng, X.; Peñaranda, S.; Verma, N.; Shulman, S.T.; Sircar, K.; Robinson, C.C.; Schmidt, T.; et al. Increased Activity of Coxsackievirus B1 Strains Associated with Severe Disease among Young Infants in the United States, 2007–2008. Clin. Infect. Dis. 2009, 49, e44–e51. [Google Scholar] [CrossRef] [PubMed]
- Villenave, R.; Wales, S.Q.; Hamkins-Indik, T.; Papafragkou, E.; Weaver, J.C.; Ferrante, T.C.; Bahinski, A.; Elkins, C.A.; Kulka, M.; Ingber, D.E. Human Gut-On-A-Chip Supports Polarized Infection of Coxsackie B1 Virus In Vitro. PLoS ONE 2017, 12, e0169412. [Google Scholar] [CrossRef] [PubMed]
- Wufuer, M.; Lee, G.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S. Skin-on-a-Chip Model Simulating Inflammation, Edema and Drug-Based Treatment. Sci. Rep. 2016, 6, 37471. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Morimoto, Y.; Takeuchi, S. Skin Integrated with Perfusable Vascular Channels on a Chip. Biomaterials 2017, 116, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.Y.; Kim, J.; Song, H.J.; Kim, K.; Choi, K.C.; Park, S.; Sung, G.Y. Development of Wrinkled Skin-on-a-Chip (WSOC) by Cyclic Uniaxial Stretching. J. Ind. Eng. Chem. 2018. [Google Scholar] [CrossRef]
- Sriram, G.; Alberti, M.; Dancik, Y.; Wu, B.; Wu, R.; Feng, Z.; Ramasamy, S.; Bigliardi, P.L.; Bigliardi-Qi, M.; Wang, Z. Full-Thickness Human Skin-on-Chip with Enhanced Epidermal Morphogenesis and Barrier Function. Mater. Today 2018, 21, 326–340. [Google Scholar] [CrossRef]
- Park, J.; Lee, B.K.; Jeong, G.S.; Hyun, J.K.; Lee, C.J.; Lee, S.-H. Three-Dimensional Brain-on-a-Chip with an Interstitial Level of Flow and Its Application as an in Vitro Model of Alzheimer’s Disease. Lab Chip 2015, 15, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Kilic, O.; Pamies, D.; Lavell, E.; Schiapparelli, P.; Feng, Y.; Hartung, T.; Bal-Price, A.; Hogberg, H.T.; Quinones-Hinojosa, A.; Guerrero-Cazares, H.; et al. Brain-on-a-Chip Model Enables Analysis of Human Neuronal Differentiation and Chemotaxis. Lab Chip 2016, 16, 4152–4162. [Google Scholar] [CrossRef] [PubMed]
- Dauth, S.; Maoz, B.M.; Sheehy, S.P.; Hemphill, M.A.; Murty, T.; Macedonia, M.K.; Greer, A.M.; Budnik, B.; Parker, K.K. Neurons Derived from Different Brain Regions Are Inherently Different in Vitro: A Novel Multiregional Brain-on-a-Chip. J. Neurophysiol. 2017, 117, 1320–1341. [Google Scholar] [CrossRef] [PubMed]
- Jastrzebska, E.; Tomecka, E.; Jesion, I. Heart-on-a-Chip Based on Stem Cell Biology. Biosens. Bioelectron. 2016, 75, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating Heart on a Chip: A Novel Microfluidic Platform to Generate Functional 3D Cardiac Microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Kobuszewska, A.; Tomecka, E.; Zukowski, K.; Jastrzebska, E.; Chudy, M.; Dybko, A.; Renaud, P.; Brzozka, Z. Heart-on-a-Chip: An Investigation of the Influence of Static and Perfusion Conditions on Cardiac (H9C2) Cell Proliferation, Morphology, and Alignment. SLAS Technol. 2017, 22, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Ardoña, H.A.M.; Lind, J.U.; Eweje, F.; Kim, S.L.; Gonzalez, G.M.; Liu, Q.; Zimmerman, J.F.; Pyrgiotakis, G.; Zhang, Z.; et al. Mussel-Inspired 3D Fiber Scaffolds for Heart-on-a-Chip Toxicity Studies of Engineered Nanomaterials. Anal. Bioanal. Chem. 2018, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Christoffersson, J.; Meier, F.; Kempf, H.; Schwanke, K.; Coffee, M.; Beilmann, M.; Zweigerdt, R.; Mandenius, C.-F. A Cardiac Cell Outgrowth Assay for Evaluating Drug Compounds Using a Cardiac Spheroid-on-a-Chip Device. Bioengineering 2018, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Kamei, K.; Kato, Y.; Hirai, Y.; Ito, S.; Satoh, J.; Oka, A.; Tsuchiya, T.; Chen, Y.; Tabata, O. Integrated Heart/Cancer on a Chip to Reproduce the Side Effects of Anti-Cancer Drugs in Vitro. RSC Adv. 2017, 7, 36777–36786. [Google Scholar] [CrossRef]
- Luni, C.; Serena, E.; Elvassore, N. Human-on-chip for therapy development and fundamental science. Curr. Opin. Biotechnol. 2014, 25, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, U.S.; Bischofs, I.B. Physical determinants of cell organization in soft media. Med. Eng. Phys. 2005, 27, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Weibel, D.B.; Whitesides, G.M. Applications of microfluidics in chemical biology. Curr. Opin. Chem. Biol. 2006, 10, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Luni, C.; Feldman, H.C.; Pozzobon, M.; De Coppi, P.; Meinhart, C.D.; Elvassore, N. Microliter-bioreactor array with buoyancy-driven stirring for human hematopoietic stem cell culture. Biomicrofluidics 2010, 4, 034105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronaldson-Bouchard, K.; Vunjak-Novakovic, G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 2018, 22, 310–324. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Zheng, W.; Zhang, W.; Jiang, X. Organs on Microfluidic Chips: A Mini Review. Sci. China Chem. 2014, 57, 356–364. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.K.; Schimek, K.; Hasenberg, T.; Ramme, A.P.; Hübner, J.; Lindner, M.; Drewell, C.; Bauer, S.; Thomas, A.; et al. A Four-Organ-Chip for Interconnected Long-Term Co-Culture of Human Intestine, Liver, Skin and Kidney Equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Abaci, H.E.; Shuler, M.L. Human-on-a-Chip Design Strategies and Principles for Physiologically Based Pharmacokinetics/Pharmacodynamics Modeling. Integr. Biol. 2015, 7, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Coppeta, J.R.; Mescher, M.J.; Isenberg, B.C.; Spencer, A.J.; Kim, E.S.; Lever, A.R.; Mulhern, T.J.; Prantil-Baun, R.; Comolli, J.C.; Borenstein, J.T. A Portable and Reconfigurable Multi-Organ Platform for Drug Development with Onboard Microfluidic Flow Control. Lab Chip 2017, 17, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Abdulwahab, S.; Choi, K.; Lafrenière, N.M.; Mudrik, J.M.; Gomaa, H.; Ahmado, H.; Behan, L.-A.; Casper, R.F.; Wheeler, A.R. A Microfluidic Technique for Quantification of Steroids in Core Needle Biopsies. Anal. Chem. 2015, 87, 4688–4695. [Google Scholar] [CrossRef] [PubMed]





| Material | Relevant Property | Proposed Application | Reference |
|---|---|---|---|
| Collagen (Chitosan) | Biocompatibility, versatile control of structure and chemistry | Bio-sensing, film assembly | [21,22] |
| Silkworm (Bombyx mori) | Biocompatibility, mechanically robust, flexibility, high mechanical modulus, and toughness | Fabrication of microfluidic channel | [23,24] |
| Agarose hydrogel | Lox cytotoxicity, biodegradability, mechanical stability at low solid fractions | Cell culture, sensors, and actuators | [25,26,27] |
| Teflon | Ease of fabrication with maximum chemical resistance | High precision assay, super clean tools, valves, and pumps fabrication | [28] |
| Acrylonitrile Butadiene Styrene (ABS) | High resolution, excellent surface finish | Making of the master mold, microfluidics interface (MI), pathogen detection, biological assay | [29,30,31,32,33,34] |
| Photocurable resin/polymer | Very high resolution with small features | Biology observation of cell growth | [35,36] |
| ABS, polycarbonate, polyphenylsulfone, elastomers | Cheap material, ease of support removal | Pathogen detection of bacteria and viruses | [37,38] |
| Polyamide | Fast build speed, multi-material printing, very durable and high-temperature stable material | Making of the master mold | [39,40] |
| Hydrogels | Swelling and contraction, act as sensors and actuators | Self-regulating valves, microlens arrays, drug release systems, binding of antigens and proteins and glucose. Flow sensors pH regulators, flooding cooling devices. | [29,41,42] |
| Polyurethane-methacrylate (PUMA) | Economical to manufacture, biocompatible, nontoxic, strong electroosmotic mobility | High-aspect-ratio microstructures | [43] |
| Polyethylene glycols (PEGs) | Relatively inexpensive, available in a wide variety of molecular weights, biocompatible, negligible cytotoxicity | Microfluidic valves, Channel cover to improve the microfluidic lifetime | [44,45] |
| Polyhydroxyalkanoates (PHAs) | Biocompatibility, tunable biodegradability | Microfilm barrier for vapor and oxygen | [46] |
| Gelatin methacrylate (gel-MA) | Photopolymerizable, porous membrane | Mechanistic vascular and valvular biology cell support matrix | [47,48] |
| Polylactic acid (PLA) and Polyglycolic acid (PGA) | Tunable biodegradation | Porous scaffold for cell culture with better adhesion | [49] |
| Poly(polyol sebacate) (PPS) | Biocompatibility, design adaptability, mechanical compliance, low cytotoxicity, degradability | 3-D microfluidic system, Microbioreactor | [50] |
| Poly(ethylene glycol) diacrylate (PEGDA) and gelatin methacryloyl (GelMA) | Biocompatibility, neovascularization potential, multi-material fabrication capability at a high spatial resolution | Tissue engineering, regenerative medicine, and bio-sensing | [51] |
| Poly(methyl methacrylate) | Favorable mechanical and thermal resistance, chemical compatibility | Genomic analysis | [52] |
| Styrene Ethylene Butylene Styrene (SEBS) | Biocompatibility, Rheological characteristics | Fabrication of complex and more sophisticated microfluidic networks (μFNs) | [53] |
| Styrene Ethylene Butylene Styrene (SEBS) | Electrical surface properties, stable and relatively high zeta potential magnitude | Microdevices for Electrokinetic Applications | [54] |
| Styrene Ethylene Butylene Styrene (SEBS) | Reduced drug absorption, Optical transmittance, Mechanical performance | Cell culture | [55] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E.; Hernández-Antonio, A.; Ahmed, I.; Sharma, A.; Parra-Saldívar, R.; Iqbal, H.M.N. Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines 2018, 9, 536. https://doi.org/10.3390/mi9100536
Sosa-Hernández JE, Villalba-Rodríguez AM, Romero-Castillo KD, Aguilar-Aguila-Isaías MA, García-Reyes IE, Hernández-Antonio A, Ahmed I, Sharma A, Parra-Saldívar R, Iqbal HMN. Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines. 2018; 9(10):536. https://doi.org/10.3390/mi9100536
Chicago/Turabian StyleSosa-Hernández, Juan Eduardo, Angel M. Villalba-Rodríguez, Kenya D. Romero-Castillo, Mauricio A. Aguilar-Aguila-Isaías, Isaac E. García-Reyes, Arturo Hernández-Antonio, Ishtiaq Ahmed, Ashutosh Sharma, Roberto Parra-Saldívar, and Hafiz M. N. Iqbal. 2018. "Organs-on-a-Chip Module: A Review from the Development and Applications Perspective" Micromachines 9, no. 10: 536. https://doi.org/10.3390/mi9100536
APA StyleSosa-Hernández, J. E., Villalba-Rodríguez, A. M., Romero-Castillo, K. D., Aguilar-Aguila-Isaías, M. A., García-Reyes, I. E., Hernández-Antonio, A., Ahmed, I., Sharma, A., Parra-Saldívar, R., & Iqbal, H. M. N. (2018). Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines, 9(10), 536. https://doi.org/10.3390/mi9100536