Cancer-Specific Biomarker hNQO1-Activatable Fluorescent Probe for Imaging Cancer Cells In Vitro and In Vivo
Abstract
:1. Introduction
2. Results
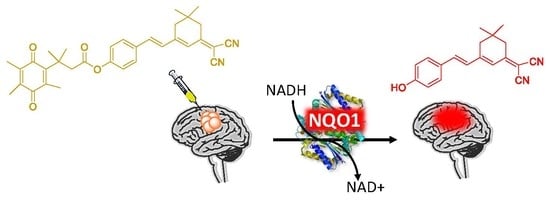
2.1. Design of the hNQO1 Responsive Fluorescent Probe
2.2. Spectroscopic Response of NQ-DCP to hNQO1
2.3. Selectivity of NQ-DCP Towards hNQO1
2.4. Visualization of Cancer Cells by Fluorescence Imaging of hNQO1
2.5. Applicability of NQ-DCP for Flow Cytometry
2.6. Evaluation of Specificity in Cancer Cells
2.7. Fluorescence Imaging of hNQO1 in U87MG Tumor-Bearing Mice
3. Discussion
4. Materials and Methods
4.1. Instruments and Materials
4.2. Spectroscopic Methods
4.3. Selectivity Evaluation
4.4. Evaluation of Inhibitor Efficiency
4.5. Cell Culture, Fluorescence Imaging, and Flow Cytometry Analysis
4.6. Knockdown of NQO1 by Small Interfering RNA (siRNA)
4.7. Immunoblotting
4.8. In Vivo Bioimaging
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Eranster, L.; Navazio, F. Soluble diaphorase in animal tissues. Acta Chem. Scand. 1958, 12, 595–602. [Google Scholar] [CrossRef]
- Li, R.; Bianchet, M.A.; Talalay, P.; Amzel, L.M. The threedimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: Mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. USA 1995, 92, 8846–8850. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Talalay, P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010, 501, 116–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Zhang, Y.; Wu, Q.; Cui, X.; Lin, Z.; Liu, S.; Chen, L. Clinical implications of high NQO1 expression in breast cancers. J. Exp. Clin. Cancer Res. 2014, 33, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, X.; Jin, T.; Wang, X.; Jin, G.; Li, Z.; Lin, L. NAD(P)H: Quinone oxidoreductase-1 overexpression predicts poor prognosis in small cell lung cancer. Oncol. Rep. 2014, 32, 2589–2595. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Jin, T.; Men, J.; Lin, Z.; Qi, P.; Piao, Y.; Yan, G. NQO1 protein expression predicts poor prognosis of non-small cell lung cancers. BMC Cancer 2015, 15, 207. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Bey, E.A.; Li, L.S.; Kabbani, W.; Yan, J.; Xie, X.J.; Hsieh, J.T.; Gao, J.; Boothman, D.A. Prostate cancer radiosensitization through PARP-1 hyperactivation. Cancer Res. 2010, 70, 8088–8096. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Qin, Y.; Jin, T.; Liu, S.; Zhang, S.; Shen, X.; Lin, Z. Significance of NQO1 overexpression for prognostic evaluation of gastric adenocarcinoma. Exp. Mol. Pathol. 2014, 96, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Wei, Y.; Jiang, T.; Wang, S. Correlation of Nrf2, NQO1, MRP1, cmyc and p53 in colorectal cancer and their relationships to clinicopathologic features and survival. Int. J. Clin. Exp. Pathol. 2014, 7, 1124–1131. [Google Scholar] [PubMed]
- Lewis, A.M.; Ough, M.; Du, J.; Tsao, M.S.; Oberley, L.W.; Cullen, J.J. Targeting NAD(P)H:Quinone Oxidoreductase (NQO1) in Pancreatic Cancer. Mol. Carcinog. 2017, 56, 1825–1834. [Google Scholar] [CrossRef] [PubMed]
- Okamura, T.; Kurisu, K.; Yamamoto, W.; Takano, H.; Nishiyama, M. NADPH/quinone oxidoreductase is a priority target of glioblastoma chemotherapy. Int. J. Oncol. 2000, 16, 295–598. [Google Scholar] [CrossRef] [PubMed]
- Li, L.S.; Reddy, S.; Lin, Z.H.; Liu, S.; Park, H.; Chun, S.G.; Bornmann, W.G.; Thibodeaux, J.; Yan, J.; Chakrabarti, G.; et al. NQO1-Mediated Tumor-Selective Lethality and Radiosensitization for Head and Neck Cancer. Mol. Cancer Ther. 2016, 15, 1757–1767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, D.; Ma, K.; Wu, X.; Hao, H.; Jiang, S. NAD(P)H:Quinone Oxidoreductase 1 (NQO1) as a Therapeutic and Diagnostic Target in Cancer. J. Med. Chem. 2018, 61, 6983–7003. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, H.M.; Kansanen, E.; Polonen, P.; Heinaniemi, M.; Levonen, A.L. Role of the Keap1-Nrf2 pathway in cancer. Adv. Cancer Res. 2014, 122, 281–320. [Google Scholar] [PubMed]
- Nioi, P.; McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: Reassessment of the ARE consensus sequence. Biochem. J. 2003, 374, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Belykh, E.; Martirosyan, N.L.; Yagmurlu, K.; Miller, E.J.; Eschbacher, J.M.; Izadyyazdanabadi, M.; Bardonova, L.A.; Byvaltsev, V.A.; Nakaji, P.; Preul, M.C. Intraoperative Fluorescence Imaging for Personalized Brain Tumor Resection: Current State and Future Directions. Front. Surg. 2016, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Stummer, W.; Reulen, H.J.; Meinel, T.; Pichlmeier, U.; Schumacher, W.; Tonn, J.C.; Rohde, V.; Oppel, F.; Turowski, B.; Woiciechowsky, C.; et al. Extent of resection and survival in glioblastoma multiforme: Identification of and adjustment for bias. Neurosurgery 2008, 62, 564–576. [Google Scholar] [PubMed]
- Zhang, D.Y.; Singhal, S.; Lee, J.Y.K. Optical Principles of Fluorescence-Guided Brain Tumor Surgery: A Practical Primer for the Neurosurgeon. Neurosurgery 2018. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.I.; Clark, P.A.; Zhang, R.R.; Kandela, I.K.; Farhoud, M.; Weichert, J.P.; Kuo, J.S. Fluorescent Cancer-Selective Alkylphosphocholine Analogs for Intraoperative Glioma Detection. Neurosurgery 2015, 76, 115–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Xie, J.; Chen, X. Activatable molecular probes for cancer imaging. Curr. Top. Med. Chem. 2010, 10, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Wirth, D.; Snuderl, M.; Sheth, S.; Kwon, C.S.; Frosch, M.P.; Curry, W.; Yaroslavsky, A.N. Identifying brain neoplasms using dye-enhanced multimodal confocal imaging. J. Biomed Opt. 2012, 17, 026012. [Google Scholar] [CrossRef] [PubMed]
- Dias, G.G.; King, A.; de Moliner, M.; Vendrell, M.; da Siva Junior, E.N. Quinone based fluorophores for imaging biological processes. Chem. Soc. Rev. 2018, 47, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.F.; Hollabaugh, N.M.; Hettiarachchi, S.U.; McCarley, R.L. Human NAD(P)H:Quinone Oxidoreductase Type I (hNQO1) Activation of Quinone Propionic Acid Trigger Groups. Biochemistry 2012, 51, 8014–8026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silvers, W.C.; Payne, A.S.; McCarley, R.L. Shedding light by cancer redox—human NAD(P)H: Quinone oxidoreductase 1 activation of a cloaked fluorescent dye. Chem. Commun. 2011, 47, 11264–11266. [Google Scholar] [CrossRef] [PubMed]
- Best, Q.A.; Johnson, A.E.; Prasai, B.; Rouillere, A.; McCarley, R.L. Environmentally Robust Rhodamine Reporters for Probe-based Cellular Detection of the Cancer-linked Oxidoreductase hNQO1. ACS Chem. Biol. 2016, 11, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Hettiarachchi, S.U.; Prasai, B.; McCarley, R.L. Detection and Cellular Imaging of Human Cancer Enzyme Using a Turn-On, Wavelength-Shiftable, Self-Immolative Profluorophore. J. Am. Chem. Soc. 2014, 136, 7575–7578. [Google Scholar] [CrossRef] [PubMed]
- Prasai, B.; Silvers, W.C.; McCarley, R.L. Oxidoreductase-Facilitated Visualization and Detection of Human Cancer Cells. Anal. Chem. 2015, 87, 6411–6418. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Luo, F.; Liu, X.; Liu, W.; Chen, W.; Liu, F.; Kuang, Y.Q.; Jiang, J.H. A novel two-photon fluorescent probe with a long Stokes shift and a high signal-to background ratio for human NAD(P)H:quinone oxidoreductase 1 (hNQO1) detection and imaging in living cells and tissues. Analyst 2017, 142, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Cuff, S.; Lewis, R.D.; Chinje, E.; Jaffar, M.; Knox, R.; Weeks, I. An improved cell-permeable fluorogenic substrate as the basis for a highly sensitive test for NAD(P)H quinone oxidoreductase 1 (NQO1) in living cells. Free Radic. Biol. Med. 2018, 116, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Zhou, L.; Wang, F.; Shi, B.; Li, C.; Wang, R.; Zhao, C. Rational construction of probes rendering ratiometric response to the cancer-specific enzyme NQO1. Dyes Pigm. 2017, 136, 846–851. [Google Scholar] [CrossRef]
- Shin, W.S.; Lee, M.G.; Verwilst, P.; Lee, J.H.; Chi, S.G.; Kim, J.S. Mitochondria-targeted aggregation induced emission theranostics: Crucial importance of in situ activation. Chem. Sci. 2016, 7, 6050–6059. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.; Cho, M.K.; Park, S.J.; Kim, D.; Nam, S.J.; Cui, L.; Kim, H.M.; Yoon, J. An efficient two-photon fluorescent probe for human NAD(P)H:quinone oxidoreductase (hNQO1) detection and imaging in tumor cells. Chem. Commun. 2017, 53, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Prasai, B.; Nakamura, Y.; Kobayashi, H.; Jackson, M.S.; McCarley, R.L. A Near-Infrared, Wavelength-Shiftable, Turn-on Fluorescent Probe for the Detection and Imaging of Cancer Tumor Cells. ACS Chem. Biol. 2017, 12, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhai, B.B.; Peng, T.; Zhong, Z.; Xu, L.; Zhang, Q.Z.; Li, L.Y.; Yi, L.; Xi, Z. Design and synthesis of near-infrared fluorescence enhancement probes for the cancer-specific enzyme hNQO1. Dyes Pigm. 2017, 143, 245–251. [Google Scholar] [CrossRef]
- Gontijo, T.B.; de Freitas, R.P.; de Lima, G.F.; de Rezende, L.C.; Pedrosa, L.F.; Silva, T.L.F.; Goulart, M.O.; Cavalcanti, B.C.; Pessoa, C.; Bruno, M.P.; et al. Novel fluorescent lapachone based BODIPY: Synthesis, computational and electrochemical aspects, and subcellular localisation of a potent antitumour hybrid quinone. Chem. Commun. 2016, 2, 13281–13284. [Google Scholar] [CrossRef] [PubMed]
- Gontijo, T.B.; de Freitas, R.P.; Emery, F.S.; Pedrosa, L.F.; Vieira Neto, J.B.; Cavalcanti, B.C.; Pessoa, C.; King, A.; de Moliner, F.; Vendrell, M.; da Silva Júnior, E.N. On the synthesis of quinone based BODIPY hybrids: New insights on antitumor activity and mechanism of action in cancer cells. Bioorg. Med. Chem. Lett. 2017, 27, 4446–4456. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Li, X.; Xu, L.; Wang, N.; Qian, X.; You, Q.; Zhang, X. Affinity-based small fluorescent probe for NAD(P)H:quinone oxidoreductase 1 (NQO1). Design, synthesis and pharmacological evaluation. Eur. J. Med. Chem. 2017, 127, 828–839. [Google Scholar] [CrossRef] [PubMed]
- Winski, S.L.; Faig, M.; Bianchet, M.A.; Siegel, D.; Swann, E.; Fung, K.; Duncan, M.W.; Moody, C.J.; Amzel, L.M.; Ross, D. Characterization of a Mechanism Based Inhibitor of NAD(P)H:Quinone Oxidoreductase 1 by Biochemical, X-ray Crystallographic, and Mass Spectrometric Approaches. Biochemistry 2001, 40, 15135–15142. [Google Scholar] [CrossRef] [PubMed]
- Punganuru, S.R.; Madala, H.R.; Mikelis, C.M.; Dixit, A.; Arutla, V.; Srivenugopal, K.S. Conception, synthesis, and characterization of a rofecoxib-combretastatin hybrid drug with potent cyclooxygenase-2 (COX-2) inhibiting and microtubule disrupting activities in colon cancer cell culture and xenograft models. Oncotarget 2018, 9, 26109–26129. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Nakamura, H.; Maeda, H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Deliv. Rev. 2011, 63, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Madala, H.R.; Punganuru, S.R.; Ali-Osman, F.; Zhang, R.; Srivenugopal, K.S. Brain- and brain tumor-penetrating disulfiram nanoparticles: Sequence of cytotoxic events and efficacy in human glioma cell lines and intracranial xenografts. Oncotarget 2017, 9, 3459–3482. [Google Scholar] [CrossRef] [PubMed]







| Fluorescent Probes for hNQO1 (Ref.) | λabs/em (Stokes-Shift) | Application | Fluorescent Probes for hNQO1 (Ref.) | λabs/em (Stokes-Shift) | Application |
|---|---|---|---|---|---|
 [25] | 485/520 (35) | In vitro |  [26] | 585/624 (39) | in vitro |
 [28] | 374/490 (116) | In vitro |  [35] | 670/710 (40) | In vitro and in vivo |
 [27] | 440/525 (85) | In vitro |  [35] | 730/800 (70) | In vitro and in vivo |
 [31] | 509/542 (33) | In vitro |  [34] | 786/798 (12) | In vitro and in vivo |
 [30] | 360/450 (90) | In vitro |  Present work | 420/465 (145) | In vitro and in vivo |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Punganuru, S.R.; Madala, H.R.; Arutla, V.; Srivenugopal, K.S. Cancer-Specific Biomarker hNQO1-Activatable Fluorescent Probe for Imaging Cancer Cells In Vitro and In Vivo. Cancers 2018, 10, 470. https://doi.org/10.3390/cancers10120470
Punganuru SR, Madala HR, Arutla V, Srivenugopal KS. Cancer-Specific Biomarker hNQO1-Activatable Fluorescent Probe for Imaging Cancer Cells In Vitro and In Vivo. Cancers. 2018; 10(12):470. https://doi.org/10.3390/cancers10120470
Chicago/Turabian StylePunganuru, Surendra Reddy, Hanumantha Rao Madala, Viswanath Arutla, and Kalkunte S. Srivenugopal. 2018. "Cancer-Specific Biomarker hNQO1-Activatable Fluorescent Probe for Imaging Cancer Cells In Vitro and In Vivo" Cancers 10, no. 12: 470. https://doi.org/10.3390/cancers10120470
APA StylePunganuru, S. R., Madala, H. R., Arutla, V., & Srivenugopal, K. S. (2018). Cancer-Specific Biomarker hNQO1-Activatable Fluorescent Probe for Imaging Cancer Cells In Vitro and In Vivo. Cancers, 10(12), 470. https://doi.org/10.3390/cancers10120470