The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer
Abstract
:1. Introduction
2. The Regulation of Transcription of MiR-483-3p
2.1. The IGF2/MiR-483 Locus
2.2. MiR-483-3p and β-Catenin
2.3. MiR-483 and the Glucose Metabolism
2.4. MiR-483 and DNA Methylation
2.5. Other Mechanisms
3. Physiological Roles of MiR-483-3p
3.1. Cell Cycle Regulation
3.2. Melatonin Synthesis
3.3. Adipocytes Differentiation and Imprinting in Newborns
3.4. Matrix Production in Eye Cells
3.5. Vascular Homeostasis
3.6. Mesodermal Differentiation
4. MiR-483-3p in Cancer
4.1. MiR-483-3p Is Often Deregulated in Cancer
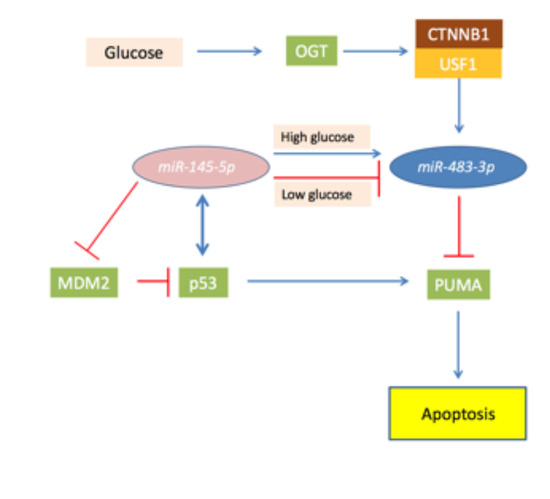
4.2. MiR-483-3p and the TP53/MiR145-5p Loop in Liver Cancer
4.3. MiR-483-3p as Cancer Biomarker
4.4. Oncosuppressor Role of MiR 483-3p
5. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNAs in cancer: Small molecules with a huge impact. J. Clin. Oncol. 2009, 27, 5848–5856. [Google Scholar] [CrossRef] [PubMed]
- Negrini, M.; Ferracin, M.; Sabbioni, S.; Croce, C.M. MicroRNAs in human cancer: From research to therapy. J. Cell Sci. 2007, 120, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Tie, Y.; Xu, C.; Zhang, Z.; Zhu, J.; Shi, Y.; Jiang, H.; Sun, Z.; Zheng, X. Identification of human fetal liver miRNAs by a novel method. FEBS Lett. 2005, 579, 3849–3854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapunzina, P. Risk of tumorigenesis in overgrowth syndromes: A comprehensive review. Am. J. Med. Genet. C Semin. Med. Genet. 2005, 137C, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, C. Igf2 and cancer. Endocr. Relat. Cancer 2013, 20, R321–R339. [Google Scholar] [CrossRef] [PubMed]
- Rainier, S.; Johnson, L.A.; Dobry, C.J.; Ping, A.J.; Grundy, P.E.; Feinberg, A.P. Relaxation of imprinted genes in human cancer. Nature 1993, 362, 747–749. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.L.; Dean, W.L.; Kelsey, G.; Allen, N.D.; Reik, W. Transactivation of IGF2 in a mouse model of beckwith-wiedemann syndrome. Nature 1997, 389, 809–815. [Google Scholar] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veronese, A.; Lupini, L.; Consiglio, J.; Visone, R.; Ferracin, M.; Fornari, F.; Zanesi, N.; Alder, H.; D’Elia, G.; Gramantieri, L.; et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 2010, 70, 3140–3149. [Google Scholar] [CrossRef] [PubMed]
- Perge, P.; Butz, H.; Pezzani, R.; Bancos, I.; Nagy, Z.; Paloczi, K.; Nyiro, G.; Decmann, A.; Pap, E.; Luconi, M.; et al. Evaluation and diagnostic potential of circulating extracellular vesicle-associated microRNAs in adrenocortical tumors. Sci. Rep. 2017, 7, 5474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Pang, W.; Zhou, Y.; Yao, W.; Xia, L.; Wang, C.; Chen, X.; Zen, K.; Zhang, C.Y.; Yuan, Y. Altered profile of serum microRNAs in pancreatic cancer-associated new-onset diabetes mellitus. J. Diabetes 2016, 8, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Peng, Q.; Lin, Y.; Zou, L.; Shen, P.; Chen, F.; Min, M.; Shen, L.; Chen, J.; Shen, B. Identification of biomarker microRNAs for predicting the response of colorectal cancer to neoadjuvant chemoradiotherapy based on microRNA regulatory network. Oncotarget 2017, 8, 2233–2248. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.; Wang, J.; Su, Q.; Chen, X.; Jiang, G.; Xu, X. Meta-analysis of the differentially expressed microRNA profiles in nasopharyngeal carcinoma. Oncotarget 2016, 7, 10513–10521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.H.; Cui, C.; Ruan, H.L.; Xue, W.Q.; Zhang, S.D.; Hu, Y.Z.; Zhou, X.X.; Jia, W.H. Plasma microRNA profiling in nasopharyngeal carcinoma patients reveals miR-548q and miR-483-5p as potential biomarkers. Chin. J. Cancer 2014, 33, 330–338. [Google Scholar] [PubMed]
- Wang, M.; Wen, T.F.; He, L.H.; Li, C.; Zhu, W.J.; Trishul, N.M. A six-microRNA set as prognostic indicators for bile duct cancer. Int. J. Clin. Exp. Med. 2015, 8, 17261–17270. [Google Scholar] [PubMed]
- Song, Q.; Xu, Y.; Yang, C.; Chen, Z.; Jia, C.; Chen, J.; Zhang, Y.; Lai, P.; Fan, X.; Zhou, X.; et al. MiR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RHOGDI1 and ALCAM. Cancer Res. 2014, 74, 3031–3042. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, Y.; Zhao, H.; Yang, X.; Luo, Y.; Ren, Y.; Liu, W.; Li, N. Serum miR-483-5p: A novel diagnostic and prognostic biomarker for patients with oral squamous cell carcinoma. Tumour Biol. 2016, 37, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Nagamitsu, Y.; Nishi, H.; Sasaki, T.; Takaesu, Y.; Terauchi, F.; Isaka, K. Profiling analysis of circulating microRNA expression in cervical cancer. Mol. Clin. Oncol. 2016, 5, 189–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer-Rochow, G.Y.; Jackson, N.E.; Conaglen, J.V.; Whittle, D.E.; Kunnimalaiyaan, M.; Chen, H.; Westin, G.; Sandgren, J.; Stalberg, P.; Khanafshar, E.; et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. Endocr. Relat. Cancer 2010, 17, 835–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, E.E.; Holloway, A.K.; Weng, J.; Fojo, T.; Kebebew, E. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer 2011, 117, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, X.; Bi, T.; Ding, Y.; Zhao, J.; Wang, C.; Jia, T.; Han, D.; Guo, G.; Wang, B.; et al. MiRNA expression signature for potentially predicting the prognosis of ovarian serous carcinoma. Tumour Biol. 2013, 34, 3501–3508. [Google Scholar] [CrossRef] [PubMed]
- Zuntini, M.; Salvatore, M.; Pedrini, E.; Parra, A.; Sgariglia, F.; Magrelli, A.; Taruscio, D.; Sangiorgi, L. MicroRNA profiling of multiple osteochondromas: Identification of disease-specific and normal cartilage signatures. Clin. Genet. 2010, 78, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Yan, L.X.; Shao, Q.; Fu, S.; Zhang, Z.C.; Ye, W.; Zeng, Y.X.; Shao, J.Y. Profiling plasma microRNA in nasopharyngeal carcinoma with deep sequencing. Clin. Chem. 2014, 60, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. Mirbase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. Mirbase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. Mirbase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. Mirbase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S. The microRNA registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef] [PubMed]
- Clokie, S.J.; Lau, P.; Kim, H.H.; Coon, S.L.; Klein, D.C. MicroRNAs in the pineal gland: MiR-483 regulates melatonin synthesis by targeting arylalkylamine N-acetyltransferase. J. Biol. Chem. 2012, 287, 25312–25324. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; French, B.A.; Li, J.; Tillman, B.; French, S.W. Altered regulation of miR-34a and miR-483-3p in alcoholic hepatitis and ddc fed mice. Exp. Mol. Pathol. 2015, 99, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Gastaldi, C.; Bourget-Ponzio, I.; Mari, B.; Meneguzzi, G.; Barbry, P.; Ponzio, G.; Rezzonico, R. Cdc25a targeting by miR-483-3p decreases ccnd-cdk4/6 assembly and contributes to cell cycle arrest. Cell Death Differ. 2013, 20, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Veronese, A.; Visone, R.; Consiglio, J.; Acunzo, M.; Lupini, L.; Kim, T.; Ferracin, M.; Lovat, F.; Miotto, E.; Balatti, V.; et al. Mutated beta-catenin evades a microRNA-dependent regulatory loop. Proc. Natl. Acad. Sci. USA 2011, 108, 4840–4845. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Liu, Y.; Jiang, J.; Liu, Y.; Yang, Z.; Wu, S.; Cao, W.; Cui, I.H.; Yu, C. Igf2-derived miR-483 mediated oncofunction by suppressing dlc-1 and associated with colorectal cancer. Oncotarget 2016, 7, 48456. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.; Sun, R.; Fu, B.; Wang, F.; Guo, C.; Tian, Z.; Wei, H. IGF-1 promotes the development and cytotoxic activity of human nk cells. Nat. Commun. 2013, 4, 1479. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Gastaldi, C.; Bourget-Ponzio, I.; Imbert, V.; Loubat, A.; Selva, E.; Busca, R.; Mari, B.; Hofman, P.; Barbry, P.; et al. MiR-483-3p controls proliferation in wounded epithelial cells. FASEB J. 2011, 25, 3092–3105. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, D.; Diekmann, U.; Fiedler, J.; Just, A.; Thum, T.; Lenzen, S.; Naujok, O. Mirnome profiling of purified endoderm and mesoderm differentiated from hescs reveals functions of miR-483-3p and miR-1263 for cell-fate decisions. Stem Cell Rep. 2017, 9, 1588–1603. [Google Scholar] [CrossRef] [PubMed]
- Arrighetti, N.; Cossa, G.; De Cecco, L.; Stucchi, S.; Carenini, N.; Corna, E.; Gandellini, P.; Zaffaroni, N.; Perego, P.; Gatti, L. Pkc-alpha modulation by miR-483-3p in platinum-resistant ovarian carcinoma cells. Toxicol. Appl. Pharmacol. 2016, 310, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Zhang, S.; Zhou, Y.; Hu, X.; Shao, C. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011, 585, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B.; Feinberg, A.P. Somatic deletion and duplication of genes on chromosome 11 in wilms’ tumours. Nature 1984, 309, 176–178. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.U.; Lidereau, R.; Theillet, C.; Callahan, R. Reduction to homozygosity of genes on chromosome 11 in human breast neoplasia. Science 1987, 238, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Feinberg, A.P.; Hamilton, S.H.; Vogelstein, B. Loss of genes on the short arm of chromosome 11 in bladder cancer. Nature 1985, 318, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Weston, A.; Willey, J.C.; Modali, R.; Sugimura, H.; McDowell, E.M.; Resau, J.; Light, B.; Haugen, A.; Mann, D.L.; Trump, B.F.; et al. Differential DNA sequence deletions from chromosomes 3, 11, 13, and 17 in squamous-cell carcinoma, large-cell carcinoma, and adenocarcinoma of the human lung. Proc. Natl. Acad. Sci. USA 1989, 86, 5099–5103. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kavanagh, J.J.; Wharton, J.T.; Wildrick, D.M.; Blick, M. Allele loss at the c-Ha-ras1 locus in human ovarian cancer. Cancer Res. 1989, 49, 1220–1222. [Google Scholar] [PubMed]
- Kurukuti, S.; Tiwari, V.K.; Tavoosidana, G.; Pugacheva, E.; Murrell, A.; Zhao, Z.; Lobanenkov, V.; Reik, W.; Ohlsson, R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to IGF2. Proc. Natl. Acad. Sci. USA 2006, 103, 10684–10689. [Google Scholar] [CrossRef] [PubMed]
- Reeve, A.E.; Eccles, M.R.; Wilkins, R.J.; Bell, G.I.; Millow, L.J. Expression of insulin-like growth factor-II transcripts in wilms’ tumour. Nature 1985, 317, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Poirier, K.; Chalas, C.; Tissier, F.; Couvert, P.; Mallet, V.; Carrie, A.; Marchio, A.; Sarli, D.; Gicquel, C.; Chaussade, S.; et al. Loss of parental-specific methylation at the IGF2 locus in human hepatocellular carcinoma. J. Pathol. 2003, 201, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Leick, M.B.; Shoff, C.J.; Wang, E.C.; Congress, J.L.; Gallicano, G.I. Loss of imprinting of IGF2 and the epigenetic progenitor model of cancer. Am. J. Stem Cells 2012, 1, 59–74. [Google Scholar] [PubMed]
- Yoshimizu, T.; Miroglio, A.; Ripoche, M.A.; Gabory, A.; Vernucci, M.; Riccio, A.; Colnot, S.; Godard, C.; Terris, B.; Jammes, H.; et al. The H19 locus acts in vivo as a tumor suppressor. Proc. Natl. Acad. Sci. USA 2008, 105, 12417–12422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Gao, F.; Tian, W.; Ruteshouser, E.C.; Wang, Y.; Lazar, A.; Stewart, J.; Strong, L.C.; Behringer, R.R.; Huff, V. Wt1 ablation and IGF2 upregulation in mice result in wilms tumors with elevated ERK1/2 phosphorylation. J. Clin. Investig. 2011, 121, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R. Cancer. Converging on beta-catenin in wilms tumor. Science 2007, 316, 988–989. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Clevers, H. Wnt signalling in stem cells and cancer. Nature 2005, 434, 843–850. [Google Scholar] [CrossRef] [PubMed]
- De La Coste, A.; Romagnolo, B.; Billuart, P.; Renard, C.A.; Buendia, M.A.; Soubrane, O.; Fabre, M.; Chelly, J.; Beldjord, C.; Kahn, A.; et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 1998, 95, 8847–8851. [Google Scholar] [CrossRef] [PubMed]
- Kahn, A. Transcriptional regulation by glucose in the liver. Biochimie 1997, 79, 113–118. [Google Scholar] [CrossRef]
- Corre, S.; Galibert, M.D. Upstream stimulating factors: Highly versatile stress-responsive transcription factors. Pigment. Cell Res. 2005, 18, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Cognard, E.; Dargaville, C.G.; Hay, D.L.; Shepherd, P.R. Identification of a pathway by which glucose regulates beta-catenin signalling via the camp/protein kinase a pathway in beta-cell models. Biochem. J. 2013, 449, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Dynkevich, Y.; Rother, K.I.; Whitford, I.; Qureshi, S.; Galiveeti, S.; Szulc, A.L.; Danoff, A.; Breen, T.L.; Kaviani, N.; Shanik, M.H.; et al. Tumors, IGF-2, and hypoglycemia: Insights from the clinic, the laboratory, and the historical archive. Endocr. Rev. 2013, 34, 798–826. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Zhao, Y.; Liu, Y.; Ma, N.; Wang, C.; Zou, J.; Liu, Z.; Zhou, Z.; Han, D.; He, J.; et al. MiR-483-3p regulates hyperglycaemia-induced cardiomyocyte apoptosis in transgenic mice. Biochem. Biophys. Res. Commun. 2016, 477, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Pepe, F.; Pagotto, S.; Soliman, S.; Rossi, C.; Lanuti, P.; Braconi, C.; Mariani-Costantini, R.; Visone, R.; Veronese, A. Regulation of miR-483-3p by the O-linked N-acetylglucosamine transferase links chemosensitivity to glucose metabolism in liver cancer cells. Oncogenesis 2017, 6, e328. [Google Scholar] [CrossRef] [PubMed]
- Slawson, C.; Housley, M.P.; Hart, G.W. O-glcnac cycling: How a single sugar post-translational modification is changing the way we think about signaling networks. J. Cell. Biochem. 2006, 97, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Hart, G.W.; Housley, M.P.; Slawson, C. Cycling of o-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007, 446, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Chen, Y.; Bian, C.; Fujiki, R.; Yu, X. Tet2 promotes histone O-glcnacylation during gene transcription. Nature 2013, 493, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ongusaha, P.P.; Miles, P.D.; Havstad, J.C.; Zhang, F.; So, W.V.; Kudlow, J.E.; Michell, R.H.; Olefsky, J.M.; Field, S.J.; et al. Phosphoinositide signalling links O-glcnac transferase to insulin resistance. Nature 2008, 451, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Olivier-Van Stichelen, S.; Guinez, C.; Mir, A.M.; Perez-Cervera, Y.; Liu, C.; Michalski, J.C.; Lefebvre, T. The hexosamine biosynthetic pathway and O-glcnacylation drive the expression of beta-catenin and cell proliferation. Am. J. Phys. Endocrinol. Metab. 2012, 302, E417–E424. [Google Scholar] [CrossRef] [PubMed]
- Olivier-Van Stichelen, S.; Dehennaut, V.; Buzy, A.; Zachayus, J.L.; Guinez, C.; Mir, A.M.; El Yazidi-Belkoura, I.; Copin, M.C.; Boureme, D.; Loyaux, D.; et al. O-glcnacylation stabilizes beta-catenin through direct competition with phosphorylation at threonine 41. FASEB J. 2014, 28, 3325–3338. [Google Scholar] [CrossRef] [PubMed]
- Shafi, R.; Iyer, S.P.; Ellies, L.G.; O’Donnell, N.; Marek, K.W.; Chui, D.; Hart, G.W.; Marth, J.D. The O-glcnac transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 2000, 97, 5735–5739. [Google Scholar] [CrossRef] [PubMed]
- Onodera, Y.; Nam, J.M.; Bissell, M.J. Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-glcnac pathways. J. Clin. Investig. 2014, 124, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Butkinaree, C.; Park, K.; Hart, G.W. O-linked beta-N-acetylglucosamine (O-glcnac): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 2010, 1800, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Comer, F.I.; Hart, G.W. Reciprocity between O-glcnac and O-phosphate on the carboxyl terminal domain of RNA polymerase ii. Biochemistry 2001, 40, 7845–7852. [Google Scholar] [CrossRef] [PubMed]
- Buren, S.; Gomes, A.L.; Teijeiro, A.; Fawal, M.A.; Yilmaz, M.; Tummala, K.S.; Perez, M.; Rodriguez-Justo, M.; Campos-Olivas, R.; Megias, D.; et al. Regulation of OGT by URI in response to glucose confers c-MYC-dependent survival mechanisms. Cancer Cell 2016, 30, 290–307. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Beatty, L.G.; Zhou, W.; Lew, J.; Schoenherr, C.; Weksberg, R.; Sadowski, P.D. Insulator and silencer sequences in the imprinted region of human chromosome 11p15.5. Hum. Mol. Genet. 2003, 12, 1927–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Maurano, M.T.; Qu, H.; Varley, K.E.; Gertz, J.; Pauli, F.; Lee, K.; Canfield, T.; Weaver, M.; Sandstrom, R.; et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome. Res. 2012, 22, 1680–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Martino, M.; Palma, G.; Azzariti, A.; Arra, C.; Fusco, A.; Esposito, F. The hmga1 pseudogene 7 induces miR-483 and miR-675 upregulation by activating EGR1 through a cerna mechanism. Genes (Basel) 2017, 8, 330. [Google Scholar] [CrossRef] [PubMed]
- Emmerling, V.V.; Fischer, S.; Stiefel, F.; Holzmann, K.; Handrick, R.; Hesse, F.; Horer, M.; Kochanek, S.; Otte, K. Temperature-sensitive miR-483 is a conserved regulator of recombinant protein and viral vector production in mammalian cells. Biotechnol. Bioeng. 2016, 113, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Emmerling, V.V.; Fischer, S.; Kleemann, M.; Handrick, R.; Kochanek, S.; Otte, K. MiR-483 is a self-regulating microRNA and can activate its own expression via USF1 in hela cells. Int. J. Biochem. Cell Biol. 2016, 80, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Bourget-Ponzio, I.; Puissant, A.; Loubat, A.; Mari, B.; Meneguzzi, G.; Auberger, P.; Barbry, P.; Ponzio, G.; Rezzonico, R. Tumor suppressor function of miR-483-3p on squamous cell carcinomas due to its pro-apoptotic properties. Cell Cycle 2013, 12, 2183–2193. [Google Scholar] [CrossRef] [PubMed]
- Ferland-McCollough, D.; Fernandez-Twinn, D.S.; Cannell, I.G.; David, H.; Warner, M.; Vaag, A.A.; Bork-Jensen, J.; Brons, C.; Gant, T.W.; Willis, A.E.; et al. Programming of adipose tissue miR-483-3p and gdf-3 expression by maternal diet in type 2 diabetes. Cell Death Differ. 2012, 19, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Bork-Jensen, J.; Thuesen, A.C.; Bang-Bertelsen, C.H.; Grunnet, L.G.; Pociot, F.; Beck-Nielsen, H.; Ozanne, S.E.; Poulsen, P.; Vaag, A. Genetic versus non-genetic regulation of miR-103, miR-143 and miR-483-3p expression in adipose tissue and their metabolic implications-a twin study. Genes 2014, 5, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, F.; Didelot, X.; Bruce, K.D.; Cagampang, F.R.; Vatish, M.; Hanson, M.; Lehnert, H.; Ceriello, A.; Byrne, C.D. Maternal high fat diet during pregnancy and lactation alters hepatic expression of insulin like growth factor-2 and key microRNAs in the adult offspring. BMC Genom. 2009, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Han, Y.; Huang, B.; Qi, Y.; Xu, L.; Guo, R.; Wang, X.; Wang, J. MicroRNA-483-3p inhibits extracellular matrix production by targeting smad4 in human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2015, 56, 8419–8427. [Google Scholar] [CrossRef] [PubMed]
- Kemp, J.R.; Unal, H.; Desnoyer, R.; Yue, H.; Bhatnagar, A.; Karnik, S.S. Angiotensin ii-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J. Mol. Cell. Cardiol. 2014, 75, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Hu, N.; Du, X.; Wang, W.; Chen, H.; Li, W.; Wei, S.; Zhuang, H.; Li, X.; Li, C. Upregulation of miR-483-3p contributes to endothelial progenitor cells dysfunction in deep vein thrombosis patients via SRF. J. Transl. Med. 2016, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Koperski, L.; Kotlarek, M.; Swierniak, M.; Kolanowska, M.; Kubiak, A.; Gornicka, B.; Jazdzewski, K.; Wojcicka, A. Next-generation sequencing reveals microRNA markers of adrenocortical tumors malignancy. Oncotarget 2017, 8, 49191–49200. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sun, Y.; Wu, H.; Zhao, D.; Chen, J. Distinguishing adrenal cortical carcinomas and adenomas: A study of clinicopathological features and biomarkers. Histopathology 2014, 64, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Guled, M.; Lahti, L.; Lindholm, P.M.; Salmenkivi, K.; Bagwan, I.; Nicholson, A.G.; Knuutila, S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma—A miRNA microarray analysis. Genes Chromosomes Cancer 2009, 48, 615–623. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.J.; Rotjanapun, K.; Hesketh, J.E.; Roy, N.C. Expression profiling indicating low selenium-sensitive microRNA levels linked to cell cycle and cell stress response pathways in the CARO-2 cell line. Br. J. Nutr. 2017, 117, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.M.; Li, J.X.; Chen, Q.; Du, H.B.; Zhang, S.Y.; Nie, J.H.; Cao, J.P.; Zhou, P.K.; Hei, T.K.; Tong, J. Radon-induced alterations in micro-RNA expression profiles in transformed BEAS2B cells. J. Toxicol. Environ. Health Part A 2013, 76, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, S.; Ragusa, M.; Urbano, F.; Filippello, A.; Di Pino, A.; Scamporrino, A.; Pulvirenti, A.; Ferro, A.; Rabuazzo, A.M.; Purrello, M.; et al. Intracellular and extracellular mirnome deregulation in cellular models of NAFLD or NASH: Clinical implications. Nutr. Metab. Cardiovasc. Dis. 2016, 26, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Chen, Y.; Feng, S.; Yi, T.; Liu, X.; Li, Q.; Liu, Z.; Zhu, C.; Hu, J.; Yu, X.; et al. MiR-483-5p promotes igf-II transcription and is associated with poor prognosis of hepatocellular carcinoma. Oncotarget 2017, 8, 99871–99888. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Cai, J.; Xu, L.; Liu, J.; Chen, M.; Zheng, M.; Wang, L.; Yang, X. MiR-483-3p regulates acute myocardial infarction by transcriptionally repressing insulin growth factor 1 expression. Mol. Med. Rep. 2018, 17, 4785–4790. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. Diana-tarbase v7.0: Indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef] [PubMed]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Li, J.; Xing, J.; Li, W.; Li, H.; Ke, X.; Zhang, J.; Ren, T.; Shang, Y.; Yang, H.; et al. Cd147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J. Hepatol. 2014, 61, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Chafey, P.; Finzi, L.; Boisgard, R.; Cauzac, M.; Clary, G.; Broussard, C.; Pegorier, J.P.; Guillonneau, F.; Mayeux, P.; Camoin, L.; et al. Proteomic analysis of beta-catenin activation in mouse liver by dige analysis identifies glucose metabolism as a new target of the wnt pathway. Proteomics 2009, 9, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Michelotti, G.A.; Machado, M.V.; Diehl, A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Pepe, F.; Lupini, L.; Ferracin, M.; Braconi, C.; Callegari, E.; Pagotto, S.; Spizzo, R.; Zagatti, B.; Lanuti, P.; Fornari, F.; et al. Over-expression of the miR-483-3p overcomes the miR-145/TP53 pro-apoptotic loop in hepatocellular carcinoma. Oncotarget 2016, 7, 31361. [Google Scholar]
- Sachdeva, M.; Mo, Y.Y. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010, 70, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Sepp-Lorenzino, L.; Prisco, M.; Linsley, P.; deAngelis, T.; Baserga, R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J. Biol. Chem. 2007, 282, 32582–32590. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Mo, Y.Y. P53 and c-MYC: How does the cell balance “yin” and “yang”? Cell Cycle 2009, 8, 1303. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.; Chow, A.S.; Au, J.S. MiR-145 inhibits cell proliferation of human lung adenocarcinoma by targeting EGFR and NUDT1. RNA Biol. 2011, 8, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, Q.; Zhang, Z.; Ge, S.; Han, Z.G.; Chen, W.T. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene 2013, 32, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zarate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzo, M.; et al. Identification by real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Ozen, M.; Creighton, C.J.; Ozdemir, M.; Ittmann, M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008, 27, 1788–1793. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sempere, L.F.; Galimberti, F.; Freemantle, S.J.; Black, C.; Dragnev, K.H.; Ma, Y.; Fiering, S.; Memoli, V.; Li, H.; et al. Uncovering growth-suppressive microRNAs in lung cancer. Clin. Cancer Res. 2009, 15, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, R.; Nicoloso, M.S.; Lupini, L.; Lu, Y.; Fogarty, J.; Rossi, S.; Zagatti, B.; Fabbri, M.; Veronese, A.; Liu, X.; et al. MiR-145 participates with tp53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2010, 17, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529–533. [Google Scholar] [CrossRef] [PubMed]
- De Queiroz, R.M.; Madan, R.; Chien, J.; Dias, W.B.; Slawson, C. Changes in O-linked N-acetylglucosamine (O-glcnac) homeostasis activate the p53 pathway in ovarian cancer cells. J. Biol. Chem. 2016, 291, 18897–18914. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yang, W.; Ma, J.; Zhang, H.; Li, Z.; Zhang, L.; Liu, J.; Han, Z.; Wang, H.; Hong, L. Role of miR-483 in digestive tract cancers: From basic research to clinical value. J. Cancer 2018, 9, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Hong, L.; Xu, G.; Hao, J.; Wang, R.; Guo, H.; Liu, J.; Zhang, Y.; Nie, Y.; Fan, D. MiR-483-3p plays an oncogenic role in esophageal squamous cell carcinoma by targeting tumor suppressor EI24. Cell Biol. Int. 2016, 40, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Abue, M.; Yokoyama, M.; Shibuya, R.; Tamai, K.; Yamaguchi, K.; Sato, I.; Tanaka, N.; Hamada, S.; Shimosegawa, T.; Sugamura, K.; et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int. J. Oncol. 2015, 46, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sun, Y.; Wu, H.; Yu, S.; Zhang, L.; Meng, Y.; Liu, M.; Yang, H.; Liu, P.; Mao, X.; et al. Elevated miR-483-3p expression is an early event and indicates poor prognosis in pancreatic ductal adenocarcinoma. Tumour Biol. 2015, 36, 9447–9456. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Wahlfors, T.; Mattila, H.; Oja, H.; Tammela, T.L.; Schleutker, J. MiRNA profiles in lymphoblastoid cell lines of finnish prostate cancer families. PLoS ONE 2015, 10, e0127427. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Qi, R.; Xu, J.; Di, Z.; Zheng, H.; Huo, W.; Zhang, L.; Chen, H.; Gao, X. Profiling of serum and urinary microRNAs in children with atopic dermatitis. PLoS ONE 2014, 9, e115448. [Google Scholar] [CrossRef] [PubMed]
- Bay, A.; Coskun, E.; Oztuzcu, S.; Ergun, S.; Yilmaz, F.; Aktekin, E. Plasma microRNA profiling of pediatric patients with immune thrombocytopenic purpura. Blood Coagul. Fibrinolysis 2014, 25, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ge, S.; Wang, X.; Yuan, Q.; Yan, Q.; Ye, H.; Che, Y.; Lin, Y.; Zhang, J.; Liu, P. Serum miR-483-5p as a potential biomarker to detect hepatocellular carcinoma. Hepatol. Int. 2013, 7, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Zhang, P.J.; Chen, W.J.; Feng, D.; Jia, Y.H.; Xie, L.X. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J. Trauma Acute Care Surg. 2012, 73, 850–854. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Nan, J.; Dong, L.; Zhang, C.; Li, H.; Na, R.; He, H.; Wang, Y. Upregulated miR-483-5p expression as a prognostic biomarker for esophageal squamous cell carcinoma. Cancer Biomark. Sect. A Dis. Markers 2017, 19, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Harling, L.; Lambert, J.; Ashrafian, H.; Darzi, A.; Gooderham, N.J.; Athanasiou, T. Elevated serum microRNA 483-5p levels may predict patients at risk of post-operative atrial fibrillation. Eur. J. Cardio-Thorac. Surg. 2017, 51, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Piao, Y.S.; Yamashita, S.; Oshima, H.; Oguma, K.; Fushida, S.; Fujimura, T.; Minamoto, T.; Seno, H.; Yamada, Y.; et al. Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene 2012, 31, 3949–3960. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, L.J.; Tan, Y.X.; Ren, H.; Qi, Z.T. MiR-138 induces cell cycle arrest by targeting cyclin d3 in hepatocellular carcinoma. Carcinogenesis 2012, 33, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, L.J.; Tan, Y.X.; Ren, H.; Qi, Z.T. Identification of deregulated miRNAs and their targets in hepatitis b virus-associated hepatocellular carcinoma. World J. Gastroenterol. 2012, 18, 5442–5453. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Wang, Q.; Wang, L.; Huang, Y.; Li, L.; Liu, L.; Zhou, X.; Xie, G.; Kang, T.; Wang, H.; et al. MiR-663, a microRNA targeting p21 (WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene 2012, 31, 4421–4433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, D.; Li, W.; Wu, X.; Gao, C.; Li, X. Identification of featured biomarkers in breast cancer with microRNA microarray. Arch. Gynecol. Obstet. 2016, 294, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ma, N.; Zhao, R.; Wu, G.; Zhang, Y.; Qiao, Y.; Han, D.; Xu, Y.; Xiang, Y.; Yan, B.; et al. Overexpression of miR-483-5p/3p cooperate to inhibit mouse liver fibrosis by suppressing the tgf-beta stimulated hscs in transgenic mice. J. Cell. Mol. Med. 2014, 18, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Milazzo, M.; Galassi, M.; Callegari, E.; Veronese, A.; Miyaaki, H.; Sabbioni, S.; Mantovani, V.; Marasco, E.; Chieco, P.; et al. p53/MDM2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol. Cancer Res. MCR 2014, 12, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Pollutri, D.; Gramantieri, L.; Bolondi, L.; Fornari, F. Tp53/microRNA interplay in hepatocellular carcinoma. Int. J. Mol. Sci. 2016, 17, 2029. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. MiR-221&222 regulate trail resistance and enhance tumorigenicity through pten and timp3 downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar] [PubMed]
- Zhang, C.Z.; Zhang, J.X.; Zhang, A.L.; Shi, Z.D.; Han, L.; Jia, Z.F.; Yang, W.D.; Wang, G.X.; Jiang, T.; You, Y.P.; et al. MiR-221 and miR-222 target puma to induce cell survival in glioblastoma. Mol. Cancer 2010, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene | Tissues | Cells/Cell Lines | References |
|---|---|---|---|
| AANAT | Pineal gland (h), (r) | Neonatal pinealocytes (r), HEK293 | [32] |
| BBC3PUMA | Kidney, Colon, Liver (h) | HEK293, HCT116, HepG2 | [12] |
| BRCA1 | Liver (m) | [33] | |
| CDC25A | Keratinocytes (h) | [34] | |
| CTNNB1 | Kidney (h), Colon (h) | HEK293, HCT116 | [35] |
| DLC1 | Colon (h) | HCT116, SW480 | [36] |
| IGF1 | Natural Killer cells (h), Cardiomyocites (m) | [37] | |
| MK2 | Keratinocyte (h) | [38] | |
| MKI67 | Keratinocyte (h) | [38] | |
| PGAM1 | Kidney (h) | [39] | |
| PRKCA | Ovarian (h) | IGROV-1 | [40] |
| SMAD4 | Pancreas (h) | SW1990, PANC1 | [41] |
| YAP1 | Keratinocyte (h) | [38] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pepe, F.; Visone, R.; Veronese, A. The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer. Cancers 2018, 10, 181. https://doi.org/10.3390/cancers10060181
Pepe F, Visone R, Veronese A. The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer. Cancers. 2018; 10(6):181. https://doi.org/10.3390/cancers10060181
Chicago/Turabian StylePepe, Felice, Rosa Visone, and Angelo Veronese. 2018. "The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer" Cancers 10, no. 6: 181. https://doi.org/10.3390/cancers10060181
APA StylePepe, F., Visone, R., & Veronese, A. (2018). The Glucose-Regulated MiR-483-3p Influences Key Signaling Pathways in Cancer. Cancers, 10(6), 181. https://doi.org/10.3390/cancers10060181