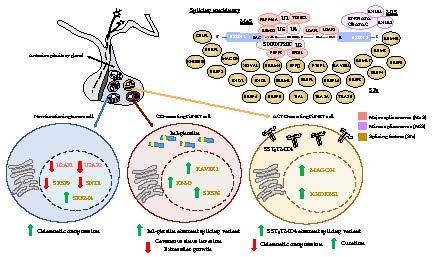
Splicing Machinery is Dysregulated in Pituitary Neuroendocrine Tumors and is Associated with Aggressiveness Features
Abstract
:1. Introduction
2. Results
2.1. Dysregulation of Splicing Machinery in NFPTs
2.2. Dysregulation of Splicing Machinery in GHomas
2.3. Dysregulation of Splicing Machinery in ACTHomas
2.4. Dysregulation of Splicing Machinery in PRLomas
2.5. Similar Dysregulation of Specific Splicing Machinery Components in all PitNET Subtypes
2.6. Effect of Pladienolide-B Treatment in PitNETs Cells
3. Discussion
4. Materials and Methods
4.1. Drugs and Reagents
4.2. Patients, Samples, and Primary Cell Cultures
4.3. Cell Lines and Culturing
4.4. RNA Extraction, Quantification and Reverse Transcription
4.5. Analysis of Splicing Machinery Components by a Customized qPCR Dynamic Array
4.6. RNA Isolation, Reverse Transcription, and Analysis of Gene Expression Levels by qPCR
4.7. Measurement of Cell Proliferation/Viability
4.8. Measurement of Hormone Secretion
4.9. Measurement of SF3B1 by Western Blotting
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mete, O.; Lopes, M.B. Overview of the 2017 WHO Classification of Pituitary Tumors. Endocr. Pathol. 2017, 28, 228–243. [Google Scholar] [CrossRef]
- Molitch, M.E. Diagnosis and Treatment of Pituitary Adenomas: A Review. JAMA 2017, 317, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Asa, S.L.; Casar-Borota, O.; Chanson, P.; Delgrange, E.; Earls, P.; Ezzat, S.; Grossman, A.; Ikeda, H.; Inoshita, N.; Karavitaki, N.; et al. From pituitary adenoma to pituitary neuroendocrine tumor (PitNET): an International Pituitary Pathology Club proposal. Endocr. Relat. Cancer 2017, 24, C5–C8. [Google Scholar] [CrossRef] [PubMed]
- Melmed, S. Pathogenesis of pituitary tumors. Nat. Rev. Endocrinol. 2011, 7, 257–266. [Google Scholar] [CrossRef]
- Caimari, F.; Korbonits, M. Novel Genetic Causes of Pituitary Adenomas. Clin. Cancer Res. 2016, 22, 5030–5042. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.J.; Reitman, Z.J.; Ma, Z.Y.; Chen, J.H.; Zhang, Q.L.; Shou, X.F.; Huang, C.X.; Wang, Y.F.; Li, S.Q.; Mao, Y.; et al. The genome-wide mutational landscape of pituitary adenomas. Cell Res. 2016, 26, 1255–1259. [Google Scholar] [CrossRef] [Green Version]
- Bi, W.L.; Horowitz, P.; Greenwald, N.F.; Abedalthagafi, M.; Agarwalla, P.K.; Gibson, W.J.; Mei, Y.; Schumacher, S.E.; Ben-David, U.; Chevalier, A.; et al. Landscape of Genomic Alterations in Pituitary Adenomas. Clin. Cancer Res. 2017, 23, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Salomon, M.P.; Wang, X.; Marzese, D.M.; Hsu, S.C.; Nelson, N.; Zhang, X.; Matsuba, C.; Takasumi, Y.; Ballesteros-Merino, C.; Fox, B.A.; et al. The Epigenomic Landscape of Pituitary Adenomas Reveals Specific Alterations and Differentiates Among Acromegaly, Cushing’s Disease and Endocrine-Inactive Subtypes. Clin. Cancer Res. 2018, 24, 4126–4136. [Google Scholar] [CrossRef]
- Zatelli, M.C. Pathogenesis of non-functioning pituitary adenomas. Pituitary 2018, 21, 130–137. [Google Scholar] [CrossRef]
- Beckers, A.; Lodish, M.B.; Trivellin, G.; Rostomyan, L.; Lee, M.; Faucz, F.R.; Yuan, B.; Choong, C.S.; Caberg, J.H.; Verrua, E.; et al. X-linked acrogigantism syndrome: Clinical profile and therapeutic responses. Endocr. Relat. Cancer 2015, 22, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.; Carlsen, E.; Marques, P.; Stiles, C.E.; Gadaleta, E.; Berney, D.M.; Roncaroli, F.; Chelala, C.; Solomou, A.; Herincs, M.; et al. Tumor microenvironment defines the invasive phenotype of AIP-mutation-positive pituitary tumors. Oncogene 2019, 38, 5381–5395. [Google Scholar] [CrossRef] [PubMed]
- Jacks, T.; Fazeli, A.; Schmitt, E.M.; Bronson, R.T.; Goodell, M.A.; Weinberg, R.A. Effects of an Rb mutation in the mouse. Nature 1992, 359, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Kiyokawa, H.; Kineman, R.D.; Manova-Todorova, K.O.; Soares, V.C.; Hoffman, E.S.; Ono, M.; Khanam, D.; Hayday, A.C.; Frohman, L.A.; Koff, A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 1996, 85, 721–732. [Google Scholar] [CrossRef]
- Ewing, I.; Pedder-Smith, S.; Franchi, G.; Ruscica, M.; Emery, M.; Vax, V.; Garcia, E.; Czirjak, S.; Hanzely, Z.; Kola, B.; et al. A mutation and expression analysis of the oncogene BRAF in pituitary adenomas. Clin. Endocrinol. 2007, 66, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kilpinen, S.; Ruusulehto, A.; Lothe, R.A.; Skotheim, R.I. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016, 35, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- Daguenet, E.; Dujardin, G.; Valcarcel, J. The pathogenicity of splicing defects: Mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015, 16, 1640–1655. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez-Costa, A.; Gahete, M.D.; Rivero-Cortes, E.; Rincon-Fernandez, D.; Nelson, R.; Beltran, M.; de la Riva, A.; Japon, M.A.; Venegas-Moreno, E.; Galvez, M.A.; et al. In1-ghrelin splicing variant is overexpressed in pituitary adenomas and increases their aggressive features. Sci. Rep. 2015, 5, 8714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luque, R.M.; Ibáñez-Costa, A.; Neto, L.V.; Taboada, G.F.; Hormaechea-Agulla, D.; Kasuki, L.; Venegas-Moreno, E.; Moreno-Carazo, A.; Gálvez, M.A.; Soto-Moreno, A.; et al. Truncated somatostatin receptor variant sst5TMD4 confers aggressive features (proliferation, invasion and reduced octreotide response) to somatotropinomas. Cancer Lett. 2015, 359, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Luque, R.M.; Sampedro-Nunez, M.; Gahete, M.D.; Ramos-Levi, A.; Ibanez-Costa, A.; Rivero-Cortes, E.; Serrano-Somavilla, A.; Adrados, M.; Culler, M.D.; Castano, J.P.; et al. In1-ghrelin, a splice variant of ghrelin gene, is associated with the evolution and aggressiveness of human neuroendocrine tumors: Evidence from clinical, cellular and molecular parameters. Oncotarget 2015, 6, 19619–19633. [Google Scholar] [CrossRef]
- Sampedro-Nunez, M.; Luque, R.M.; Ramos-Levi, A.M.; Gahete, M.D.; Serrano-Somavilla, A.; Villa-Osaba, A.; Adrados, M.; Ibanez-Costa, A.; Martin-Perez, E.; Culler, M.D.; et al. Presence of sst5TMD4, a truncated splice variant of the somatostatin receptor subtype 5, is associated to features of increased aggressiveness in pancreatic neuroendocrine tumors. Oncotarget 2016, 7, 6593–6608. [Google Scholar] [CrossRef]
- Hormaechea-Agulla, D.; Gahete, M.D.; Jimenez-Vacas, J.M.; Gomez-Gomez, E.; Ibanez-Costa, A.; Fernando, L.; Rivero-Cortes, E.; Sarmento-Cabral, A.; Valero-Rosa, J.; Carrasco-Valiente, J.; et al. The oncogenic role of the In1-ghrelin splicing variant in prostate cancer aggressiveness. Mol. Cancer 2017, 16, 146. [Google Scholar] [CrossRef] [PubMed]
- Hormaechea-Agulla, D.; Jimenez-Vacas, J.M.; Gomez-Gomez, E.; Fernando, L.; Carrasco-Valiente, J.; Valero-Rosa, J.; Moreno, M.M.; Sanchez-Sanchez, R.; Ortega-Salas, R.; Gracia-Navarro, F.; et al. The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. FASEB J. 2017, 31, 4682–4696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelechano, V.; Wei, W.; Steinmetz, L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013, 497, 127–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoskins, A.A.; Moore, M.J. The spliceosome: A flexible, reversible macromolecular machine. Trends Biochem. Sci. 2012, 37, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Wan, R.; Shi, Y. Molecular Mechanisms of pre-mRNA Splicing through Structural Biology of the Spliceosome. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef]
- Kornblihtt, A.R.; Schor, I.E.; Allo, M.; Dujardin, G.; Petrillo, E.; Munoz, M.J. Alternative splicing: A pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013, 14, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Cereda, M.; Pozzoli, U.; Rot, G.; Juvan, P.; Schweitzer, A.; Clark, T.; Ule, J. RNAmotifs: prediction of multivalent RNA motifs that control alternative splicing. Genome Biol. 2014, 15, R20. [Google Scholar] [CrossRef]
- Duran-Prado, M.; Gahete, M.D.; Hergueta-Redondo, M.; Martinez-Fuentes, A.J.; Cordoba-Chacon, J.; Palacios, J.; Gracia-Navarro, F.; Moreno-Bueno, G.; Malagon, M.M.; Luque, R.M.; et al. The new truncated somatostatin receptor variant sst5TMD4 is associated to poor prognosis in breast cancer and increases malignancy in MCF-7 cells. Oncogene 2012, 31, 2049–2061. [Google Scholar] [CrossRef] [PubMed]
- Molè, D.; Gentilin, E.; Ibáñez-Costa, A.; Gagliano, T.; Gahete, M.D.; Tagliati, F.; Rossi, R.; Pelizzo, M.R.; Pansini, G.; Luque, R.M.; et al. The expression of the truncated isoform of somatostatin receptor subtype 5 associates with aggressiveness in medullary thyroid carcinoma cells. Endocrine 2015, 50, 442–452. [Google Scholar] [CrossRef]
- Puig-Domingo, M.; Luque, R.M.; Reverter, J.L.; Lopez-Sanchez, L.M.; Gahete, M.D.; Culler, M.D.; Diaz-Soto, G.; Lomena, F.; Squarcia, M.; Mate, J.L.; et al. The truncated isoform of somatostatin receptor5 (sst5TMD4) is associated with poorly differentiated thyroid cancer. PLoS ONE 2014, 9, e85527. [Google Scholar] [CrossRef] [PubMed]
- Van Alphen, R.J.; Wiemer, E.A.; Burger, H.; Eskens, F.A. The spliceosome as target for anticancer treatment. Br. J. Cancer 2009, 100, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Di, C.; Yan, J.; Wang, F.; Qu, T.; Wang, Y.; Chen, Y.; Zhang, X.; Liu, Y.; Yang, H.; et al. Inhibition of SF3b1 by pladienolide B evokes cycle arrest, apoptosis induction and p73 splicing in human cervical carcinoma cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1273–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez-Vacas, J.M.; Herrero-Aguayo, V.; Gómez-Gómez, E.; León-González, A.J.; Sáez-Martínez, P.; Alors-Perez, E.; Fuentes-Fayos, A.C.; Martínez-López, A.; Sánchez-Sánchez, R.; González-Serrano, T.; et al. Spliceosome Component SF3B1 as Novel Prognostic Biomarker and Therapeutic Target for Prostate Cancer. Transl. Res. J. Lab. Clin. Med. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Altenberger, T.; Bilban, M.; Auer, M.; Knosp, E.; Wolfsberger, S.; Gartner, W.; Mineva, I.; Zielinski, C.; Wagner, L.; Luger, A. Identification of DLK1 variants in pituitary- and neuroendocrine tumors. Biochem. Biophys. Res. Commun. 2006, 340, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Barabutis, N.; Siejka, A.; Schally, A.V.; Block, N.L.; Cai, R.; Varga, J.L. Activation of mitogen-activated protein kinases by a splice variant of GHRH receptor. J. Mol. Endocrinol. 2010, 44, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Vitale, L.; Lenzi, L.; Huntsman, S.A.; Canaider, S.; Frabetti, F.; Casadei, R.; Facchin, F.; Carinci, P.; Zannotti, M.; Coppola, D.; et al. Differential expression of alternatively spliced mRNA forms of the insulin-like growth factor 1 receptor in human neuroendocrine tumors. Oncol. Rep. 2006, 15, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Lekva, T.; Berg, J.P.; Lyle, R.; Heck, A.; Bollerslev, J.; Ueland, T. Alternative splicing of placental lactogen (CSH2) in somatotroph pituitary adenomas. Neuro Endocrinol. Lett. 2015, 36, 136–142. [Google Scholar]
- Gahete, M.D.; Rincón-Fernández, D.; Durán-Prado, M.; Hergueta-Redondo, M.; Ibáñez-Costa, A.; Rojo-Sebastián, A.; Gracia-Navarro, F.; Culler, M.D.; Casanovas, O.; Moreno-Bueno, G.; et al. The truncated somatostatin receptor sst5TMD4 stimulates the angiogenic process and is associated to lymphatic metastasis and disease-free survival in breast cancer patients. Oncotarget 2016, 7, 60110–60122. [Google Scholar] [CrossRef] [Green Version]
- Durán-Prado, M.; Gahete, M.D.; Martinez-Fuentes, A.J.; Luque, R.M.; Quintero, A.; Webb, S.M.; Benito-Lopez, P.; Leal, A.; Schulz, S.; Gracia-Navarro, F.; et al. Identification and characterization of two novel truncated but functional isoforms of the somatostatin receptor subtype 5 differentially present in pituitary tumors. J. Clin. Endocrinol. Metab. 2009, 94, 2634–2643. [Google Scholar] [CrossRef]
- Lee, S.C.; Abdel-Wahab, O. Therapeutic targeting of splicing in cancer. Nat. Med. 2016, 22, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Ibanez-Costa, A.; Rivero-Cortes, E.; Vazquez-Borrego, M.C.; Gahete, M.D.; Jimenez-Reina, L.; Venegas-Moreno, E.; de la Riva, A.; Arraez, M.A.; Gonzalez-Molero, I.; Schmid, H.A.; et al. Octreotide and pasireotide (dis)similarly inhibit pituitary tumor cells in vitro. J. Endocrinol. 2016, 231, 135–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zawada, N.B.; Kunert-Radek, J.; Pawlikowski, M.; Pisarek, H.; Radek, M. An evaluation of the effects of somatostatin analogue therapy in non-functioning pituitary adenomas in comparison to acromegaly. Endokrynol. Pol. 2016, 67, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Taboada, G.F.; Luque, R.M.; Bastos, W.; Guimaraes, R.F.; Marcondes, J.B.; Chimelli, L.M.; Fontes, R.; Mata, P.J.; Filho, P.N.; Carvalho, D.P.; et al. Quantitative analysis of somatostatin receptor subtype (SSTR1-5) gene expression levels in somatotropinomas and non-functioning pituitary adenomas. Eur. J. Endocrinol. 2007, 156, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lupp, A.; Mendoza, N.; Martin, N.; Beschorner, R.; Honegger, J.; Schlegel, J.; Shively, T.; Pulz, E.; Schulz, S.; et al. SSTR3 is a putative target for the medical treatment of gonadotroph adenomas of the pituitary. Endocr. Relat. Cancer 2015, 22, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Wang, X.; Long, Y.; Desiderio, D.M. Heterogeneity analysis of the proteomes in clinically nonfunctional pituitary adenomas. BMC Med. Genom. 2014, 7, 69. [Google Scholar] [CrossRef]
- Fu, Y.; Huang, B.; Shi, Z.; Han, J.; Wang, Y.; Huangfu, J.; Wu, W. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing beta-catenin biosynthesis. EMBO Mol. Med. 2013, 5, 737–750. [Google Scholar] [CrossRef]
- Yoshino, H.; Enokida, H.; Chiyomaru, T.; Tatarano, S.; Hidaka, H.; Yamasaki, T.; Gotannda, T.; Tachiwada, T.; Nohata, N.; Yamane, T.; et al. Tumor suppressive microRNA-1 mediated novel apoptosis pathways through direct inhibition of splicing factor serine/arginine-rich 9 (SRSF9/SRp30c) in bladder cancer. Biochem. Biophys. Res. Commun. 2012, 417, 588–593. [Google Scholar] [CrossRef]
- Yu, L.; Xu, J.; Liu, J.; Zhang, H.; Sun, C.; Wang, Q.; Shi, C.; Zhou, X.; Hua, D.; Luo, W.; et al. The novel chromatin architectural regulator SND1 promotes glioma proliferation and invasion and predicts the prognosis of patients. Neuro Oncol. 2019, 21, 742–754. [Google Scholar] [CrossRef]
- Gu, X.; Xue, J.; Ai, L.; Sun, L.; Zhu, X.; Wang, Y.; Liu, C. SND1 expression in breast cancer tumors is associated with poor prognosis. Ann. N. Y. Acad. Sci. 2018, 1433, 53–60. [Google Scholar] [CrossRef]
- Cui, X.; Zhao, C.; Yao, X.; Qian, B.; Su, C.; Ren, Y.; Yao, Z.; Gao, X.; Yang, J. SND1 acts as an anti-apoptotic factor via regulating the expression of lncRNA UCA1 in hepatocellular carcinoma. RNA Biol. 2018, 15, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, C.; Jun, Y.; Lee, S.; Jung, Y.; Kim, J. Integrative Profiling of Alternative Splicing Induced by U2AF1 S34F Mutation in Lung Adenocarcinoma Reveals a Mechanistic Link to Mitotic Stress. Mol. Cells 2018, 41, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Choudhary, G.S.; Pellagatti, A.; Choi, K.; Bolanos, L.C.; Bhagat, T.D.; Gordon-Mitchell, S.; Von Ahrens, D.; Pradhan, K.; Steeples, V.; et al. U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol. 2019, 21, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Glasser, E.; Agrawal, A.A.; Jenkins, J.L.; Kielkopf, C.L. Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs. Biochemistry 2017, 56, 4757–4761. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, D.; Zhu, M.; Yu, H.; Pan, Z.; Liu, L.; Geng, Q.; Pan, H.; Yan, M.; Yao, M. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics 2019, 9, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Shimojo, M.; Kasahara, Y.; Inoue, M.; Tsunoda, S.I.; Shudo, Y.; Kurata, T.; Obika, S. A gapmer antisense oligonucleotide targeting SRRM4 is a novel therapeutic medicine for lung cancer. Sci. Rep. 2019, 9, 7618. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Lovnicki, J.; Chen, R.; Fazli, L.; Wang, Y.; Gleave, M.; Huang, J.; Dong, X. SRRM4 gene expression correlates with neuroendocrine prostate cancer. Prostate 2019, 79, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Del Rio-Moreno, M.; Alors-Perez, E.; Gonzalez-Rubio, S.; Ferrin, G.; Reyes, O.; Rodriguez-Peralvarez, M.; Sanchez-Frias, M.E.; Sanchez-Sanchez, R.; Ventura, S.; Lopez-Miranda, J.; et al. Dysregulation of the splicing machinery is associated to the development of non-alcoholic fatty liver disease. J. Clin. Endocrinol. Metab. 2019, 104, 3389–3402. [Google Scholar] [CrossRef]
- Gahete, M.D.; Del Rio-Moreno, M.; Camargo, A.; Alcala-Diaz, J.F.; Alors-Perez, E.; Delgado-Lista, J.; Reyes, O.; Ventura, S.; Perez-Martinez, P.; Castano, J.P.; et al. Changes in Splicing Machinery Components Influence, Precede, and Early Predict the Development of Type 2 Diabetes: From the CORDIOPREV Study. EBioMedicine 2018, 37, 356–365. [Google Scholar] [CrossRef] [Green Version]
- Melling, N.; Bachmann, K.; Hofmann, B.; El Gammal, A.T.; Reeh, M.; Mann, O.; Moebius, C.; Blessmann, M.; Izbicki, J.R.; Grupp, K. Prevalence and clinical significance of RBM3 immunostaining in non-small cell lung cancers. J. Cancer Res. Clin. Oncol. 2019, 145, 873–879. [Google Scholar] [CrossRef]
- Kang, S.H.; Cho, J.; Jeong, H.; Kwon, S.Y. High RNA-binding Motif Protein 3 Expression Is Associated with Improved Clinical Outcomes in Invasive Breast Cancer. J. Breast Cancer 2018, 21, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Grupp, K.; Hofmann, B.; Kutup, A.; Bachmann, K.; Bogoevski, D.; Melling, N.; Uzunoglu, F.G.; El Gammal, A.T.; Koop, C.; Simon, R.; et al. Reduced RBM3 expression is associated with aggressive tumor features in esophageal cancer but not significantly linked to patient outcome. BMC Cancer 2018, 18, 1106. [Google Scholar] [CrossRef] [PubMed]
- Siesing, C.; Sorbye, H.; Dragomir, A.; Pfeiffer, P.; Qvortrup, C.; Ponten, F.; Jirstrom, K.; Glimelius, B.; Eberhard, J. High RBM3 expression is associated with an improved survival and oxaliplatin response in patients with metastatic colorectal cancer. PLoS ONE 2017, 12, e0182512. [Google Scholar] [CrossRef] [PubMed]
- Casar-Borota, O.; Heck, A.; Schulz, S.; Nesland, J.M.; Ramm-Pettersen, J.; Lekva, T.; Alafuzoff, I.; Bollerslev, J. Expression of SSTR2a, but not of SSTRs 1, 3, or 5 in somatotroph adenomas assessed by monoclonal antibodies was reduced by octreotide and correlated with the acute and long-term effects of octreotide. J. Clin. Endocrinol. Metab. 2013, 98, E1730–E1739. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Wang, Q.; Yang, Q.; Wang, H.; Qiang, F.; He, S.; Cai, J.; Yang, L.; Wang, Y. Clinical significance and effect of Sam68 expression in gastric cancer. Oncol. Lett. 2018, 15, 4745–4752. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Che, H.; Li, M.; Li, X. Sam68 is Overexpressed in Epithelial Ovarian Cancer and Promotes Tumor Cell Proliferation. Med. Sci. Monit. 2016, 22, 3248–3256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stricker, T.P.; Brown, C.D.; Bandlamudi, C.; McNerney, M.; Kittler, R.; Montoya, V.; Peterson, A.; Grossman, R.; White, K.P. Robust stratification of breast cancer subtypes using differential patterns of transcript isoform expression. PLoS Genet. 2017, 13, e1006589. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Krainer, A.R. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014, 12, 1195–1204. [Google Scholar] [CrossRef]
- Kramer, A.; Gruter, P.; Groning, K.; Kastner, B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol. 1999, 145, 1355–1368. [Google Scholar] [CrossRef]
- Yokoi, A.; Kotake, Y.; Takahashi, K.; Kadowaki, T.; Matsumoto, Y.; Minoshima, Y.; Sugi, N.H.; Sagane, K.; Hamaguchi, M.; Iwata, M.; et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011, 278, 4870–4880. [Google Scholar] [CrossRef]
- Sato, M.; Muguruma, N.; Nakagawa, T.; Okamoto, K.; Kimura, T.; Kitamura, S.; Yano, H.; Sannomiya, K.; Goji, T.; Miyamoto, H.; et al. High antitumor activity of pladienolide B and its derivative in gastric cancer. Cancer Sci. 2014, 105, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, M.K.; Kumar, D.; Villa, R.; La Clair, J.J.; Benner, C.; Sasik, R.; Jones, H.; Ghia, E.M.; Rassenti, L.Z.; Kipps, T.J.; et al. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica 2015, 100, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Suda, K.; Rozeboom, L.; Yu, H.; Ellison, K.; Rivard, C.J.; Mitsudomi, T.; Hirsch, F.R. Potential effect of spliceosome inhibition in small cell lung cancer irrespective of the MYC status. PLoS ONE 2017, 12, e0172209. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Borrego, M.C.; Fuentes-Fayos, A.C.; Herrera-Martinez, A.D.; L-López, F.; Ibáñez-Costa, A.; Moreno-Moreno, P.; Alhambra-Exposito, M.R.; Barrera-Martín, A.; Blanco-Acevedo, C.; Dios, E.; et al. Biguanides exert antitumoral actions in pituitary tumor cells through AMPK-dependent and -independent mechanisms. J. Clin. Endocrinol. Metab. 2019, 104, 3501–3513. [Google Scholar] [CrossRef] [PubMed]
- Ibanez-Costa, A.; Lopez-Sanchez, L.M.; Gahete, M.D.; Rivero-Cortes, E.; Vazquez-Borrego, M.C.; Galvez, M.A.; de la Riva, A.; Venegas-Moreno, E.; Jimenez-Reina, L.; Moreno-Carazo, A.; et al. BIM-23A760 influences key functional endpoints in pituitary adenomas and normal pituitaries: molecular mechanisms underlying the differential response in adenomas. Sci. Rep. 2017, 7, 42002. [Google Scholar] [CrossRef] [PubMed]
- Luque, R.M.; Ibáñez-Costa, A.; López-Sánchez, L.M.; Jimenez-Reina, L.; Venegas-Moreno, E.; Galvez, M.A.; Villa-Osaba, A.; Madrazo-Atutxa, A.M.; Japon, M.A.; de la Riva, A.; et al. A cellular and molecular basis for the selective desmopressin-induced ACTH release in Cushing disease patients: key role of AVPR1b receptor and potential therapeutic implications. J. Clin. Endocrinol. Metab. 2013, 98, 4160–4169. [Google Scholar] [CrossRef] [PubMed]
- Uphoff, C.C.; Drexler, H.G. Detection of mycoplasma contaminations. Methods Mol. Biol. 2013, 946, 1–13. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Neto, L.V.; Machado Ede, O.; Luque, R.M.; Taboada, G.F.; Marcondes, J.B.; Chimelli, L.M.; Quintella, L.P.; Niemeyer, P., Jr.; de Carvalho, D.P.; Kineman, R.D.; et al. Expression analysis of dopamine receptor subtypes in normal human pituitaries, nonfunctioning pituitary adenomas and somatotropinomas, and the association between dopamine and somatostatin receptors with clinical response to octreotide-LAR in acromegaly. J. Clin. Endocrinol. Metab. 2009, 94, 1931–1937. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–141091. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Borrego, M.C.; Fuentes-Fayos, A.C.; Venegas-Moreno, E.; Rivero-Cortés, E.; Dios, E.; Moreno-Moreno, P.; Madrazo-Atutxa, A.; Remón, P.; Solivera, J.; Wildemberg, L.E.; et al. Splicing Machinery is Dysregulated in Pituitary Neuroendocrine Tumors and is Associated with Aggressiveness Features. Cancers 2019, 11, 1439. https://doi.org/10.3390/cancers11101439
Vázquez-Borrego MC, Fuentes-Fayos AC, Venegas-Moreno E, Rivero-Cortés E, Dios E, Moreno-Moreno P, Madrazo-Atutxa A, Remón P, Solivera J, Wildemberg LE, et al. Splicing Machinery is Dysregulated in Pituitary Neuroendocrine Tumors and is Associated with Aggressiveness Features. Cancers. 2019; 11(10):1439. https://doi.org/10.3390/cancers11101439
Chicago/Turabian StyleVázquez-Borrego, Mari C., Antonio C. Fuentes-Fayos, Eva Venegas-Moreno, Esther Rivero-Cortés, Elena Dios, Paloma Moreno-Moreno, Ainara Madrazo-Atutxa, Pablo Remón, Juan Solivera, Luiz E. Wildemberg, and et al. 2019. "Splicing Machinery is Dysregulated in Pituitary Neuroendocrine Tumors and is Associated with Aggressiveness Features" Cancers 11, no. 10: 1439. https://doi.org/10.3390/cancers11101439
APA StyleVázquez-Borrego, M. C., Fuentes-Fayos, A. C., Venegas-Moreno, E., Rivero-Cortés, E., Dios, E., Moreno-Moreno, P., Madrazo-Atutxa, A., Remón, P., Solivera, J., Wildemberg, L. E., Kasuki, L., López-Fernández, J. M., Gadelha, M. R., Gálvez-Moreno, M. A., Soto-Moreno, A., Gahete, M. D., Castaño, J. P., & Luque, a. R. M. (2019). Splicing Machinery is Dysregulated in Pituitary Neuroendocrine Tumors and is Associated with Aggressiveness Features. Cancers, 11(10), 1439. https://doi.org/10.3390/cancers11101439