SerpinB3 Differently Up-Regulates Hypoxia Inducible Factors -1α and -2α in Hepatocellular Carcinoma: Mechanisms Revealing Novel Potential Therapeutic Targets
Abstract
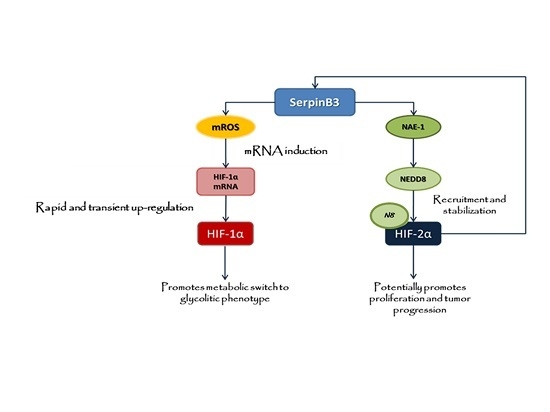
:1. Introduction
2. Results
2.1. SerpinB3 Overexpression Up-Regulates HIF-1α and HIF-2α Hepatic Protein Levels
2.2. Up-Regulation of HIF-1α by SerpinB3 Is Related to Intracellular Generation of ROS
2.3. SerpinB3 Induces HIF-2α Stabilization through Selective NEDDylation
2.4. SerpinB3 Up-Regulated HIF-α Subunits Are Biologically Active
2.5. SerpinB3 Was Able to Modulate Angiogenesis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Cell Lines and Culture Conditions
4.2.2. Mouse Models
4.2.3. Patients and Samples
4.2.4. Quantitative Real-Time PCR (qPCR)
4.2.5. NAE1 and HIFs Silencing by Small RNA Interference
- NAE1: 5′-AAGTAGTGTGTTAATGATTGA-3′
- HIF-2α: 5′-CCCGGATAGACTTATTGCCAA-3′
- HIF-1α: 5′-AGGAAGAACTATGAACATAAA-3′
4.2.6. Western Blot Analysis
4.2.7. Immunohistochemistry Analysis
4.2.8. Detection of Intracellular Generation of ROS
4.2.9. Lactic Acid Release in Culture Medium
4.2.10. Measurements of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR)
4.2.11. Proteasome Activity Assay
4.2.12. Cell Cycle Analysis
4.2.13. Soft Agar Colony Formation
4.2.14. In Vitro Angiogenesis
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pontisso, P.; Calabrese, F.; Benvegnù, L.; Lise, M.; Belluco, C.; Ruvoletto, M.G.; Marino, M.; Valente, M.; Nitti, D.; Gatta, A.; et al. Overexpression of squamous cell carcinoma antigen variants in hepatocellular carcinoma. Br. J. Cancer 2004, 90, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Kato, H. Expression and function of squamous cell carcinoma antigen. Anticancer Res. 1996, 16, 2149–2153. [Google Scholar] [PubMed]
- Guido, M.; Roskams, T.; Pontisso, P.; Fassan, M.; Thung, S.N.; Giacomelli, L.; Sergio, A.; Farinati, F.; Cillo, U.; Rugge, M. Squamous cell carcinoma antigen in human liver carcinogenesis. J. Clin. Pathol. 2008, 61, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Turato, C.; Calabrese, F.; Biasiolo, A.; Quarta, S.; Ruvoletto, M.; Tono, N.; Paccagnella, D.; Fassina, G.; Merkel, C.; Harrison, T.J.; et al. SERPINB3 modulates TGF-β expression in chronic liver disease. Lab. Investig. 2010, 90, 1016–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turato, C.; Buendia, M.A.; Fabre, M.; Redon, M.J.; Branchereau, S.; Quarta, S.; Ruvoletto, M.; Perilongo, G.; Grotzer, M.A.; Gatta, A.; et al. Over-expression of SERPINB3 in hepatoblastoma: A possible insight into the genesis of this tumour? Eur. J. Cancer 2012, 48, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Beneduce, L.; Ph, D.; Castaldi, F.; Ph, D.; Marino, M.; Quarta, S.; Ph, D.; Ruvoletto, M.; Gatta, A.; Pontisso, P.; et al. Squamous Cell Carcinoma Antigen-Immunoglobulin M Complexes as Novel Biomarkers for Hepatocellular Carcinoma. Cancer 2005, 103, 2558–2565. [Google Scholar] [CrossRef]
- Vidalino, L.; Doria, A.; Quarta, S.; Zen, M.; Gatta, A.; Pontisso, P. SERPINB3, apoptosis and autoimmunity. Autoimmun. Rev. 2009, 9, 108–112. [Google Scholar] [CrossRef]
- Villano, G.; Quarta, S.; Ruvoletto, M.G.; Turato, C.; Vidalino, L.; Biasiolo, A.; Tono, N.; Lunardi, F.; Calabrese, F.; Dall’olmo, L.; et al. Role of squamous cell carcinoma antigen-1 on liver cells after partial hepatectomy in transgenic mice. Int. J. Mol. Med. 2010, 25, 137–143. [Google Scholar]
- Quarta, S.; Vidalino, L.; Turato, C.; Ruvoletto, M.; Calabrese, F.; Valente, M.; Cannito, S.; Fassina, G.; Parola, M.; Gatta, A.; et al. SERPINB3 induces epithelial - Mesenchymal transition. J. Pathol. 2010, 221, 343–356. [Google Scholar] [CrossRef]
- Sheshadri, N.; Catanzaro, J.M.; Bott, A.J.; Sun, Y.; Ullman, E.; Chen, E.I.; Pan, J.-A.; Wu, S.; Crawford, H.C.; Zhang, J.; et al. SCCA1/SERPINB3 Promotes Oncogenesis and Epithelial-Mesenchymal Transition via the Unfolded Protein Response and IL6 Signaling. Cancer Res. 2014, 74, 6318–6329. [Google Scholar] [CrossRef] [Green Version]
- Catanzaro, J.M.; Sheshadri, N.; Zong, W.-X. SerpinB3/B4: Mediators of Ras-driven inflammation and oncogenesis. Cell Cycle 2014, 13, 3155–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turato, C.; Cannito, S.; Simonato, D.; Villano, G.; Morello, E.; Terrin, L.; Quarta, S.; Biasiolo, A.; Ruvoletto, M.; Martini, A.; et al. SerpinB3 and Yap Interplay Increases Myc Oncogenic Activity. Sci Rep. 2015, 5, 17701. [Google Scholar] [CrossRef] [PubMed]
- Cannito, S.; Turato, C.; Paternostro, C.; Biasiolo, A.; Colombatto, S.; Cambieri, I.; Quarta, S.; Novo, E.; Morello, E.; Villano, G.; et al. Hypoxia up-regulates SERPINB3 through HIF-2α in human liver cancer cells. Oncotarget 2015, 6, 2206–2221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brahimi-Horn, M.C.; Chiche, J.; Pouysségur, J. Hypoxia and cancer. J. Mol. Med. 2007, 85, 1301–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rankin, E.B.; Giaccia, A.J. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008, 15, 678–685. [Google Scholar] [CrossRef] [Green Version]
- Cannito, S.; Novo, E.; Compagnone, A.; di Bonzo, L.V.; Busletta, C.; Zamara, E.; Paternostro, C.; Povero, D.; Bandino, A.; Bozzo, F.; et al. Redox mechanisms switch on hypoxia-dependent epithelial-mesenchymal transition in cancer cells. Carcinogenesis 2008, 29, 2267–2278. [Google Scholar] [CrossRef]
- Hill, R.P.; Marie-egyptienne, D.T.; Hedley, D.W. Cancer Stem Cells, Hypoxia and Metastasis. Semin. Radiat. Oncol. 2009, 19, 106–111. [Google Scholar] [CrossRef]
- Borovski, T.; De Sousa E Melo, F.; Vermeulen, L.; Medema, J.P. Cancer Stem Cell Niche: The Place to Be. Cancer Res. 2011, 71, 634–639. [Google Scholar] [CrossRef] [Green Version]
- Koh, M.Y.; Lemos, R.; Liu, X.; Powis, G. The hypoxia-associated factor switches cells from HIF-1α- to HIF-2α-dependent signaling promoting stem cell characteristics, aggressive tumor growth and invasion. Cancer Res. 2011, 71, 4015–4027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niecknig, H.; Tug, S.; Delos Reyes, B.; Kirsch, M. Role of reactive oxygen species in the regulation of HIF-1 by prolyl hydroxylase 2 under mild hypoxia. Free Radic. Res. 2012, 466, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Movafagh, S.; Crook, S.; Vo, K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: New developments in an old debate. J. Cell. Biochem. 2015, 116, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Kietzmann, T.; Görlach, A. Reactive oxygen species in the control of hypoxia-inducible factor-mediated gene expression. Semin. Cell Dev. Biol. 2005, 16, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Choi, A.M.K. Heme Oxygenase-1: Redox Regulation of a Stress Protein in Lung and Cell Culture Models. Antioxid Redox Signal. 2005, 7, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; Jiang, B.H.; Chin, B.Y.; Iyer, N.V.; Alam, J.; Semenza, G.L.; Choi, A.M. Hypoxia-inducible factor-1 mediates transcriptional activation of the heme oxygenase-1 gene in response to hypoxia. J. Biol. Chem. 1997, 272, 5375–5381. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Wang, L.; Chodosh, L.A.; Keith, B.; Simon, M.C. Differential Roles of Hypoxia-Inducible Factor 1 alpha ( HIF-1 alpha ) and HIF-2 alpha in Hypoxic Gene Regulation. Mol. Cell. Biol. 2003, 23, 9361–9374. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, D.; Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007, 315, 201–205. [Google Scholar] [CrossRef] [Green Version]
- Miranda, M.; Sorkin, A. Regulation of receptors and transporters by ubiquitination: New insights into surprisingly similar mechanisms. Mol. Interv. 2007, 7, 157–167. [Google Scholar] [CrossRef]
- Abidi, N.; Xirodimas, D.P. Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic. Endocr. Relat. Cancer 2015, 22, T55–T70. [Google Scholar] [CrossRef] [Green Version]
- Barbier-Torres, L.; Delgado, T.C.; García-Rodríguez, J.L.; Zubiete-Franco, I.; Fernández-Ramos, D.; Buqué, X.; Cano, A.; Gutiérrez-de Juan, V.; Fernández-Domínguez, I.; Lopitz-Otsoa, F.; et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget 2015, 6, 2509–2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leidecker, O.; Matic, I.; Mahata, B.; Pion, E.; Xirodimas, D.P. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 2012, 11, 1142–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maghames, C.M.; Lobato-gil, S.; Perrin, A.; Trauchessec, H.; Rodriguez, M.S.; Urbach, S.; Marin, P.; Xirodimas, D.P. NEDDylation promotes nuclear protein aggregation upon proteotoxic stress. Nat. Commun. 2018, 9, 4376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef]
- Luo, Z.; Yu, G.; Lee, H.W.; Li, L.; Wang, L.; Yang, D.; Pan, Y.; Ding, C.; Qian, J.; Wu, L.; et al. The Nedd8-Activating Enzyme Inhibitor MLN4924 Induces Autophagy and Apoptosis to Suppress Liver Cancer Cell Growth. Cancer Res. 2012, 72, 3360–3372. [Google Scholar] [CrossRef] [Green Version]
- Keith, B.; Johnson, R.S.; Simon, M.C. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 2012, 12, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Turato, C.; Vitale, A.; Fasolato, S.; Ruvoletto, M.; Terrin, L.; Quarta, S.; Morales, R.R.; Biasiolo, A. SERPINB3 is associated with TGF- b 1 and cytoplasmic b -catenin expression in hepatocellular carcinomas with poor prognosis. Br. J. Cancer 2014, 110, 2708–2715. [Google Scholar] [CrossRef] [Green Version]
- Courtney, K.D.; Infante, J.R.; Lam, E.T.; Figlin, R.A.; Rini, B.I.; Brugarolas, J.; Zojwalla, N.J.; Lowe, A.M.; Wang, K.; Wallace, E.M.; et al. Phase I dose-escalation trial of PT2385, a first-in-class hypoxia-inducible factor-2a antagonist in patients with previously treated advanced clear cell renal cell carcinoma. J. Clin. Oncol. 2018, 36, 867–874. [Google Scholar] [CrossRef]
- Xie, C.; Gao, X.; Sun, D.; Zhang, Y.; Krausz, K.W.; Qin, X.; Gonzalez, F.J. Metabolic profiling of the novel hypoxia-inducible factor 2α inhibitor PT2385 in vivo and in vitro. Drug Metab. Dispos. 2018, 46, 336–345. [Google Scholar] [CrossRef] [Green Version]
- Guzy, R.D.; Hoyos, B.; Robin, E.; Chen, H.; Liu, L.; Mansfield, K.D.; Simon, M.C.; Hammerling, U.; Schumacker, P.T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005, 1, 401–408. [Google Scholar] [CrossRef] [Green Version]
- Sharp, F.R.; Bernaudin, M. HIF1 and oxygen sensing in the brain. Nat. Rev. Neurosci. 2004, 5, 437–448. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Luo, Q.; Li, L.; Jia, L. Neddylation: A novel modulator of the tumor microenvironment. Mol. Cancer 2019, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Embade, N.; Fernández-ramos, D.; Varela-rey, M.; Beraza, N.; Juan, V.G.D.; Woodhoo, A.; Martínez-lópez, N.; Bustamante, F.J.; Belén, A.; Hoz, D.; et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 2012, 55, 1237–1248. [Google Scholar] [CrossRef] [Green Version]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G., Jr. HIFα Targeted for VHL-Mediated Destruction by Proline Hydroxylation: Implications for O2 Sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Curtis, V.F.; Ehrentraut, S.F.; Campbell, E.L.; Glover, L.E.; Bayless, A.; Kelly, C.J.; Kominsky, D.J.; Colgan, S.P. Stabilization of HIF through inhibition of Cullin-2 neddylation is protective in mucosal inflammatory responses. FASEB J. 2015, 29, 208–215. [Google Scholar] [CrossRef] [Green Version]
- Russell, R.C.; Ohh, M. NEDD8 acts as a “molecular switch” defining the functional selectivity of VHL. EMBO Rep. 2008, 9, 486–491. [Google Scholar] [CrossRef]
- Ryu, J.-H.; Li, S.-H.; Park, H.-S.; Park, J.-W.; Lee, B.; Chun, Y.-S. Hypoxia-inducible factor α subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J. Biol. Chem. 2011, 286, 6963–6970. [Google Scholar] [CrossRef] [Green Version]
- Maranchie, J.K.; Vasselli, J.R.; Riss, J.; Bonifacino, J.S.; Linehan, W.M.; Klausner, R.D. The contribution of VHL substrate binding and HIF1-α to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 2002, 1, 247–255. [Google Scholar] [CrossRef] [Green Version]
- Kondo, K.; Klco, J.; Nakamura, E.; Lechpammer, M.; Kaelin, W.G. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 2002, 1, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Koshiji, M.; Kageyama, Y.; Pete, E.A.; Horikawa, I.; Barrett, J.C.; Huang, L.E. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004, 23, 1949–1956. [Google Scholar] [CrossRef] [Green Version]
- Novo, E.; Villano, G.; Turato, C.; Cannito, S.; Paternostro, C.; Busletta, C.; Biasiolo, A.; Quarta, S.; Morello, E.; Bocca, C.; et al. SerpinB3 Promotes Pro-fibrogenic Responses in Activated Hepatic Stellate Cells. Sci. Rep. 2017, 7, 3420. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Kesarwala, A.H.; Eggert, T.; Medina-Echeverz, J.; Kleiner, D.E.; Jin, P.; Stroncek, D.F.; Terabe, M.; Kapoor, V.; ElGindi, M.; et al. NAFLD causes selective CD4(+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature 2016, 531, 253–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novo, E.; Busletta, C.; Bonzo, L.V.D.; Povero, D.; Paternostro, C.; Mareschi, K.; Ferrero, I.; David, E.; Bertolani, C.; Caligiuri, A.; et al. Intracellular reactive oxygen species are required for directional migration of resident and bone marrow-derived hepatic pro-fibrogenic cells. J. Hepatol. 2011, 54, 964–974. [Google Scholar] [CrossRef] [PubMed]
- Morello, E.; Sutti, S.; Foglia, B.; Novo, E.; Cannito, S.; Bocca, C.; Rajsky, M.; Bruzzì, S.; Abate, M.L.; Rosso, C.; et al. Hypoxia-inducible factor 2α drives nonalcoholic fatty liver progression by triggering hepatocyte release of histidine-rich glycoprotein. Hepatology 2018, 67, 2196–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbier-Torres, L.; Beraza, N.; Fernández-Tussy, P.; Lopitz-Otsoa, F.; Fernández-Ramos, D.; Zubiete-Franco, I.; Varela-Rey, M.; Delgado, T.C.; Gutiérrez, V.; Anguita, J.; et al. Histone Deacetylase 4 promotes cholestatic liver injury in the absence of Prohibitin-1 HHS Public Access. Hepatology 2015, 62, 1237–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cascone, I.; Giraudo, E.; Caccavari, F.; Napione, L.; Bertotti, E.; Collard, J.G.; Serini, G.; Bussolino, F. Temporal and spatial modulation of Rho GTPases during in vitro formation of capillary vascular network. Adherens junctions and myosin light chain as targets of Rac1 and RhoA. J. Biol. Chem. 2003, 278, 50702–50713. [Google Scholar] [PubMed] [Green Version]
- Zhou, L.; Zhang, W.; Sun, Y.; Jia, L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell. Signal. 2018, 44, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Martínez-chantar, M.L.; Fern, D. NEDDylation in liver cancer: The regulation of the RNA binding protein Hu antigen R. Pancreatology 2015, 15 (Suppl. 4), S49–S54. [Google Scholar]
- Swords, R.T.; Kelly, K.R.; Smith, P.G.; Garnsey, J.J.; Mahalingam, D.; Medina, E.; Oberheu, K.; Padmanabhan, S.; O’Dwyer, M.; Nawrocki, S.T.; et al. Inhibition of NEDD8-activating enzyme: A novel approach for the treatment of acute myeloid leukemia. Blood 2010, 115, 3796–3800. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhang, J.; Lin, X.; Wu, D.; Li, G.; Zhong, C.; Fang, L.; Jiang, P.; Yin, L.; Zhang, L.; et al. Inhibition of neddylation modification by MLN4924 sensitizes hepatocellular carcinoma cells to sorafenib. Oncol. Rep. 2019, 41, 3257–3269. [Google Scholar] [CrossRef]
- Bangoura, G.; Liu, Z.-S.; Qian, Q.; Jiang, C.-Q.; Yang, G.-F.; Jing, S. Prognostic significance of HIF-2α/EPAS1 expression in hepatocellular carcinoma. World J. Gastroenterol. 2007, 13, 3176–3182. [Google Scholar] [CrossRef] [PubMed]












| Primer | Sense | Reverse |
| Human HK4 | 5’-CGCCAAGAAGGAGAAGGTAGAGC-3’ | 5’-AAGTCCCCGACTTCTGAGCC-3’ |
| Human PKM2 | 5’-GCTGTCTGGAGAAACAGCCA-3’ | 5’-GTGACTTGAGGCTCGCACAA-3’ |
| Human HK2 | 5’-AAATGGAGCGAGGTCTGAGC-3’ | 5’-CCCACTTCCCATTCCGAACA-3’ |
| Human PDK3 | 5′-ATTCAATGCCAAAGCGCCAG-3′ | AGCAGGGTAGCCCTCTTTT-3′ |
| Human LDHA | 5′-CATGGCCTGTGCCATCAGTA-3′ | GAAAAGGCTGCCATGTTGG-3′ |
| Human EPO | 5′-GAGCCCAGAAGGAAGCCATC-3′ | CGGAAAGTGTCAGCAGTGA-3 |
| Humman CXCR4 | 5′-TCCATTCCTTTGCCTCTTTTGC-3′ | CGGAAACAGGGTTCCTTCAT-3′ |
| Human MMP9 | 5’-TTCAGGGAGACGCCCATTTC-3’ | AAACCGAGTTGGAACCACGA-3’ |
| Human NAE-1 | 5’-GCAAACAAAAACAAATGAAGCCAG-3’ | 5’-TTTCAGTTCCTGTGGCTGTTG-3’ |
| Murine NAE-1 | 5’-AGCGGAGAAGATGCTGGAAAC-3’ | 5’-CTGGACTCTCTTCCACAAAACT-3’ |
| Murine HIF-1α | 5’-TCAAGTCAGCAACGTGGAAG-3’ | 5’-TATCGAGGCTGTGTCGACTG-3’ |
| Murine HIF-2α | 5’-CTAAGTGGCCTGTGGGTGAT-3’ | 5’-GTGTCTTGGAAGGCTTGCTC-3’ |
| Human HIF-1α | 5’-CCACCTATGACCTGCTTGGT-3’ | 5’-TATCCAGGCTGTGTCGACTG-3’ |
| Human HIF-2α | 5’-TTGATGTGGAAACGGATGAA-3’ | 5’-GGAACCTGCTCTTGCTGTTC-3’ |
| Human HO-1 | 5’-ATGACACCAAGGACCAGAGC-3’ | 5’-GTGTAAGGACCCATCGGAGA-3’ |
| Human GAPDH | 5′-TGGTATCGTGGAAGGACTCATGAC-3′ | 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′ |
| Murine GAPDH | 5′-TGGAAAGCTGTGGCGTGAT-3′ | 5′-TGCTTCACCACCTTCTTGAT-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cannito, S.; Foglia, B.; Villano, G.; Turato, C.; Delgado, T.C.; Morello, E.; Pin, F.; Novo, E.; Napione, L.; Quarta, S.; et al. SerpinB3 Differently Up-Regulates Hypoxia Inducible Factors -1α and -2α in Hepatocellular Carcinoma: Mechanisms Revealing Novel Potential Therapeutic Targets. Cancers 2019, 11, 1933. https://doi.org/10.3390/cancers11121933
Cannito S, Foglia B, Villano G, Turato C, Delgado TC, Morello E, Pin F, Novo E, Napione L, Quarta S, et al. SerpinB3 Differently Up-Regulates Hypoxia Inducible Factors -1α and -2α in Hepatocellular Carcinoma: Mechanisms Revealing Novel Potential Therapeutic Targets. Cancers. 2019; 11(12):1933. https://doi.org/10.3390/cancers11121933
Chicago/Turabian StyleCannito, Stefania, Beatrice Foglia, Gianmarco Villano, Cristian Turato, Teresa C Delgado, Elisabetta Morello, Fabrizio Pin, Erica Novo, Lucia Napione, Santina Quarta, and et al. 2019. "SerpinB3 Differently Up-Regulates Hypoxia Inducible Factors -1α and -2α in Hepatocellular Carcinoma: Mechanisms Revealing Novel Potential Therapeutic Targets" Cancers 11, no. 12: 1933. https://doi.org/10.3390/cancers11121933
APA StyleCannito, S., Foglia, B., Villano, G., Turato, C., Delgado, T. C., Morello, E., Pin, F., Novo, E., Napione, L., Quarta, S., Ruvoletto, M., Fasolato, S., Zanus, G., Colombatto, S., Lopitz-Otsoa, F., Fernández-Ramos, D., Bussolino, F., Sutti, S., Albano, E., ... Parola, M. (2019). SerpinB3 Differently Up-Regulates Hypoxia Inducible Factors -1α and -2α in Hepatocellular Carcinoma: Mechanisms Revealing Novel Potential Therapeutic Targets. Cancers, 11(12), 1933. https://doi.org/10.3390/cancers11121933