Competing Endogenous RNA Networks in the Epithelial to Mesenchymal Transition in Diffuse-Type of Gastric Cancer
Abstract
:Simple Summary
Abstract
1. Introduction

2. The Molecular Pathology of the Epithelial-Mesenchymal Transition
2.1. E-CADHERIN
2.2. ZEB
2.3. SNAIL and SLUG
2.4. TWIST
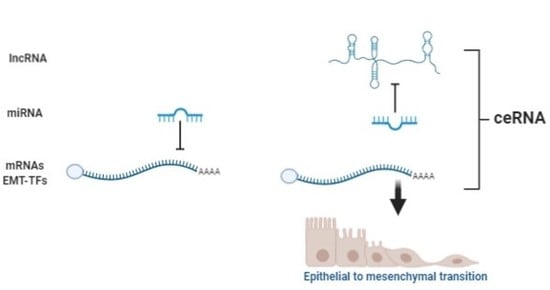
3. Noncoding RNAs: Classification and Functions
3.1. ncRNA in the EMT Process
3.2. miRNA, lncRNA, and ceRNA as Direct Regulators of E-Cadherin
| miRNA | mRNA | Function | Ref. |
|---|---|---|---|
| Upregulation miRNAs | |||
| miR-9 | E-cadherin | Upregulation promotes EMT through the direct binding of E-cadherin | [64] |
| miR-199a | E-cadherin | Upregulation promotes EMT through the direct binding of E-cadherin | [66] |
| miR-217 | E-cadherin | Upregulation promotes EMT through the direct binding of E-cadherin | [67] |
| miR-544a | E-cadherin | Upregulation promotes EMT through the direct binding of E-cadherin | [68] |
| Downregulation miRNAs | |||
| miR-141 | ZEB1/2 | Downregulation promotes EMT by increasing ZEB1 and ZEB 2 | [69] |
| miR-200c | ZEB1/2 | Downregulation promotes EMT by increasing ZEB1 and ZEB 2 | [69] |
| miR-200c | ZEB1/2 | Downregulation promotes EMT by increasing ZEB1 and ZEB 2 | [70] |
| miR-205-3p | ZEB1/2 | Downregulation promotes EMT by increasing ZEB1 and ZEB2 | [71] |
| miR-574-3p | ZEB1 | Downregulation promotes EMT by increasingZEB1 | [72] |
| miR-145 | ZEB2 | Downregulation promotes EMT by increasing ZEB2 | [73] |
| miR-200b | ZEB2 | Downregulation promotes EMT by increasing ZEB2 | [74] |
| miR-338-3p | ZEB2 | Downregulation promotes EMT by increasing ZEB2 | [75] |
| miR-506 | ZEB2 | Downregulation promotes EMT by increasing ZEB2 | [76] |
| miR-22 | Snail | Downregulation promotes EMT by increasing Snail | [77] |
| miR-153 | Snail | Downregulation promotes EMT by increasing Snail | [78] |
| miR-195 | Snail | Downregulation promotes EMT by increasing Snail | [79] |
| miR-204 | Snail | Downregulation promotes EMT by increasing Snail | [80] |
| miR-491 | Snail | Downregulation promotes EMT by increasing Snail | [81] |
| miR-33a | Slug | Downregulation promotes EMT by increasing Slug | [82] |
| miR-124 | Slug | Downregulation promotes EMT by increasing Slug | [83] |
| miR-203 | Slug | Downregulation promotes EMT by increasing Slug | [84] |
| miR-506 | Slug | Downregulation promotes EMT by increasing Slug | [85] |
| miR-15a-3p | Twist | Downregulation promotes EMT by increasing Twist | [86] |
| miR-16-1-3p | Twist | Downregulation promotes EMT by increasing Twist | [86] |
| miR-186 | Twist | Downregulation promotes EMT by increasing Twist | [87] |
| miR-381 | Twist | Downregulation promotes EMT by increasing Twist | [88] |
| miR-495 | Twist | Downregulation promotes EMT by increasing Twist | [89] |
| lncRNA | mRNA | Function | Ref. |
|---|---|---|---|
| Upregulated lncRNAs | |||
| AFAP1-AS1 | E-cadherin | Upregulation of AFAP1-AS1 decreases E-cadherin through unknown mechanism and promotes EMT | [104] |
| AGAP2-AS1 | E-cadherin | Upregulation of AGAP2-AS1 decreases E-cadherin through unknown mechanism and promotes EMT | [90] |
| AOC4P | E-cadherin | Upregulation of AOC4P decreases E-cadherin through unknown mechanism and promotes EMT | [98] |
| CCAT1 | E-cadherin | Upregulation of CCAT1 decreases E-cadherin through unknown mechanism and promotes EMT | [92] |
| LINC00941 | E-cadherin | Upregulation of LINC00941 decreases E-cadherin through unknown mechanism and promotes EMT | [99] |
| LNC01614 | E-cadherin | Upregulation of LNC01614 decreases E-cadherin through unknown mechanism and promotes EMT | [97] |
| LOC554202 | E-cadherin | Upregulation of LOC554202 decreases E-cadherin through unknown mechanism and promotes EMT | [105] |
| MNX1-AS1 | E-cadherin | Upregulation of MNX1-AS1 decreases E-cadherin through unknown mechanism and promotes EMT | [94] |
| NEAT1 | E-cadherin | Upregulation of NEAT1 decreases E-cadherin through unknown mechanism and promotes EMT | [95] |
| SNHG20 | E-cadherin | Upregulation of SNHG20 decreases E-cadherin through unknown mechanism and promotes EMT | [93] |
| TPT1-AS1 | E-cadherin | Upregulation of TPT1-AS1 decreases E-cadherin through unknown mechanism and promotes EMT | [91] |
| DNM3OS | Snail | Upregulation of DNM3OS promotes EMT through unknown mechanism and increases Snail | [106] |
| HOTAIR | Snail | Upregulation of HOTAIR promotes EMT through unknown mechanism and increases Snail | [107] |
| MALAT1 | Snail | Upregulation of MALAT1 promotes EMT through unknown mechanism and increases Snail | [108] |
| XLOC_010235 | Snail | Upregulation of XLOC_010235 promotes EMT through unknown mechanism and increases Snail | [109] |
| LINC00978 | Slug | Upregulation of LINC00978 promotes EMT through unknown mechanism and increases Slug | [110] |
| Linc-GPR65-1 | Slug | Upregulation of Linc-GPR65-1 promotes EMT through unknown mechanism and increases Slug | [111] |
| FRLnc1 | Twist | Upregulation of FRLnc1 promotes EMT through unknown mechanism and increases Twist | [112] |
| ZFAS1 | Twist | Upregulation of ZFAS1 promotes EMT through unknown mechanism and increases Twist | [113] |
| TUG1 | EZH2 | Upregulation of TUG1 promotes development of GC by increasing EZH2 | [100] |
| downregulated lncRNAs | |||
| MEG3 | E-cadherin | Downregulation of MEG3 decrease E-cadherin through unknown mechanism and promotes EMT | [101] |
| LINC00261 | Slug | Downregulation of LINC00261 prevents the degradation of Slug favoring EMT | [114] |
| lncRNA | State | miRNA | State | mRNA | State | Function | Ref. |
|---|---|---|---|---|---|---|---|
| RP11-789C1.1 | Down | miR-5003-3p | Up | E-cadherin | Down | RP11-789C1.1/miR-5003-3p/E-cadherin axis promotes EMT | [103] |
| CASC15 | Up | miR-33a | Down | ZEB1 | Up | CASC15/miR-33a/ZEB1 axis promotes EMT | [115] |
| CAT104 | Up | miR-381 | Down | ZEB1 | Up | CAT104/miR-381/ZEB1 axis promotes EMT | [116] |
| H19 | Up | miR-141 | Down | ZEB1 | Up | H19/miR-141/ZEB1 axis promotes EMT | [117] |
| MAGI2-AS3 | Up | miR-141-3p | Down | ZEB1 | Up | MAGI2-AS3/miR-141-3p/ZEB1 axis promotes EMT | [118] |
| MAGI2-AS3 | Up | miR-200a-3p | Down | ZEB1 | Up | MAGI2-AS3/200a-3p/ZEB1 axis promotes EMT | [118] |
| SNHG6 | Up | miR-101-3p | Down | ZEB1 | Up | SNHG6/miR-101-3p/ZEB1 axis promotes EMT | [119] |
| ZEB1-AS1 | Up | miR-149-3p | Down | ZEB1 | Up | ZEB1-AS1/miR-149-3p/ZEB1 axis promotes EMT | [120] |
| UCA1 | Up | miR-203 | Down | ZEB2 | Up | UCA1/miR-203/ZEB2 axis promotes EMT | [121] |
| GCMA | Up | miR-34a | Down | Snail | Up | GCMA/miR-34a/SNAIL axis promotes EMT | [122] |
| GCMA | Up | miR-124 | Down | Snail | Up | GCMA/miR-124/SNAIL axis promotes EMT | [122] |
| H19 | Up | miR-22-3p | Down | Snail | Up | H19/miR-22-3p/Snail1 axis promotes EMT | [123] |
| PVT1 | Up | miR-30a | Down | Snail | Up | PVT1/miR-30a/SNAIL axis promotes EMT | [124] |
| SNHG7 | Up | miR-34a | Down | Snail | Up | SNHG7/miR-34a/SNAIL axis promotes EMT | [125] |
| XIST | Up | miR-101 | Down | EZH2 | Up | XIST/miR-101/EZH2 axis promotes EMT | [126] |
| lncRNA-ATB | Up | miR-141-3p | Down | TGF-β2 | Up | lnc-ATB/miR-141-3p/TGF-β2 axis promotes EMT | [127] |
3.3. miRNA, lncRNA, and ceRNA in the Regulation of the EMT-Inducing Transcription Factors
3.3.1. ZEB
3.3.2. SNAIL
3.3.3. SLUG
3.3.4. TWIST
4. Prediction of ceRNAs for EMT Processes
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.E.; Strong, V.E. Gastric Cancer Etiology and Management in Asia and the West. Annu. Rev. Med. 2019, 70, 353–367. [Google Scholar] [CrossRef]
- Bonequi, P.; Meneses-González, F.; Correa, P.; Rabkin, C.S.; Camargo, M.C. Risk factors for gastric cancer in Latin America: A meta-analysis. Cancer Causes Control 2012, 24, 217–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abnet, C.C.; Corley, U.A.; Freedman, N.D.; Kamangar, F. Diet and upper gastrointestinal malignancies. Gastroenterol 2015, 148, 1234–1243.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Wei, J.; He, X.; An, P.; Wang, H.; Jiang, L.; Shao, D.; Liang, H.; Li, Y.; Wang, F.; et al. Landscape of dietary factors associated with risk of gastric cancer: A systematic review and dose-response meta-analysis of prospective cohort studies. Eur. J. Cancer 2015, 51, 2820–2832. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, J.; Cheng, A.S.L.; Yu, J.; To, K.-F.; Kang, W. Gastric cancer: Genome damaged by bugs. Oncogene 2020, 39, 3427–3442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slavin, T.P.; Weitzel, J.N.; Neuhausen, S.L.; Schrader, K.A.; Oliveira, C.; Karam, R. Genetics of gastric cancer: What do we know about the genetic risks? Transl. Gastroenterol. Hepatol. 2019, 4, 55. [Google Scholar] [CrossRef]
- Carrasco-Avino, G.; Corvalán, A.H. Helicobacter pylori-Induced Chronic Gastritis and Assessing Risks for Gastric Cancer. Gastroenterol. Res. Pr. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Laurén, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Mariette, C.; European Chapter of International Gastric Cancer Association; Carneiro, F.; Grabsch, H.I.; Van Der Post, R.S.; Allum, W.; De Manzoni, G. Consensus on the pathological definition and classification of poorly cohesive gastric carcinoma. Gastric Cancer 2018, 22, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Kim, J.-H.; Chun, J.; Yoon, Y.H.; Park, H. Chronic atrophic gastritis and intestinal metaplasia surrounding diffuse-type gastric cancer: Are they just bystanders in the process of carcinogenesis? PLoS ONE 2019, 14, e0226427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henson, D.E.; Dittus, C.; Younes, M.; Nguyen, H.; Albores-Saavedra, J. Differential trends in the intestinal and diffuse types of gastric carcinoma in the United States, 1973-2000: Increase in the signet ring cell type. Arch. Pathol. Lab. Med. 2004, 128, 765–770. [Google Scholar]
- Howson, C.P.; Hiyama, T.; Wynder, E.L. The decline in gastric cancer: Epidemiology of an unplanned triumph. Epidemiol. Rev. 1986, 8, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Faria, G.; Pinto-De-Sousa, J.; Preto, J.R.; Santos-Sousa, H.; Barbosa, J.A.; Costa-Maia, J. Three decades of clinical-pathological trends in gastric cancer: Prospective data from a Portuguese hospital. Int. J. Surg. 2013, 11, 472–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, P.J.; on behalf of the AGAMENON study group; Carmona-Bayonas, A.; Hernández, R.; Custodio, A.; Cano, J.M.; LaCalle, A.; Echavarria, I.; Macias, I.; Mangas, M.; et al. Lauren subtypes of advanced gastric cancer influence survival and response to chemotherapy: Real-world data from the AGAMENON National Cancer Registry. Br. J. Cancer 2017, 117, 775–782. [Google Scholar] [CrossRef] [Green Version]
- Pattison, S.; Mitchell, C.; Lade, S.; Leong, T.; Busuttil, R.A.; Boussioutas, A. Early relapses after adjuvant chemotherapy suggests primary chemoresistance in diffuse gastric cancer. PLoS ONE 2017, 12, e0183891. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, E.; Busuttil, R.A.; Kong, J.C.; Pattison, S.; Sung, J.J.Y.; Yu, J.; El-Omar, E.M.; Simpson, J.A.; Boussioutas, A. A cohort study and meta-analysis of the evidence for consideration of Lauren subtype when prescribing adjuvant or palliative chemotherapy for gastric cancer. Ther. Adv. Med. Oncol. 2020, 12. [Google Scholar] [CrossRef]
- Chuaqui, B.; Chuaqui, R.; Duarte, I.; González, S.; Etchart, M.; Rosenberg, H. Lecciones de Anatomía Patológica; Pontificia Universidad Católica de Chile: Santiago, Chile, 1996. [Google Scholar]
- Ge, S.; Xia, X.; Ding, C.; Zhen, B.; Zhou, Q.; Feng, J.; Yuan, J.; Chen, R.; Li, Y.; Ge, Z.; et al. A proteomic landscape of diffuse-type gastric cancer. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Blair, V.R.; McLeod, M.; Carneiro, F.; Coit, D.G.; D’Addario, J.L.; Van Dieren, J.M.; Harris, K.L.; Hoogerbrugge, N.; Oliveira, C.; Van Der Post, R.S.; et al. Hereditary diffuse gastric cancer: Updated clinical practice guidelines. Lancet Oncol. 2020, 21, e386–e397. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Yi, B.-R.; Kim, N.-H.; Choi, K.-C. Role of the epithelial-mesenchymal transition and its effects on embryonic stem cells. Exp. Mol. Med. 2014, 46, e108. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Weinberg, R.A. Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Front. Med. 2018, 12, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Bruner, H.C.; Derksen, P.W.B. Loss of E-Cadherin-Dependent Cell–Cell Adhesion and the Development and Progression of Cancer. Cold Spring Harb. Perspect. Biol. 2017, 10, a029330. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Ballestar, E.; Esteller, M.; Cano, A. Snail Mediates E-Cadherin Repression by the Recruitment of the Sin3A/Histone Deacetylase 1 (HDAC1)/HDAC2 Complex. Mol. Cell. Biol. 2004, 24, 306–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puisieux, A.; Brabletz, T.; Caramel, J. Oncogenic roles of EMT-inducing transcription factors. Nature 2014, 16, 488–494. [Google Scholar] [CrossRef]
- Imani, S.; Hosseinifard, H.; Cheng, J.; Wei, C.; Fu, J. Prognostic Value of EMT-inducing Transcription Factors (EMT-TFs) in Metastatic Breast Cancer: A Systematic Review and Meta-analysis. Sci. Rep. 2016, 6, 28587. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Abascal, M.F.; Besso, M.J.; Rosso, M.; Mencucci, M.V.; Aparicio, E.; Szapiro, G.; I Furlong, L.; Vazquez-Levin, M.H. CDH1/E-cadherin and solid tumors. An updated gene-disease association analysis using bioinformatics tools. Comput. Biol. Chem. 2016, 60, 9–20. [Google Scholar] [CrossRef]
- Mendonsa, A.M.; Na, T.-Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef]
- Gall, T.M.H.; E Frampton, A. Gene of the month: E-cadherin (CDH1). J. Clin. Pathol. 2013, 66, 928–932. [Google Scholar] [CrossRef] [PubMed]
- Koplev, S.; Lin, K.; Dohlman, A.B.; Ma’Ayan, A. Integration of pan-cancer transcriptomics with RPPA proteomics reveals mechanisms of epithelial-mesenchymal transition. PLoS Comput. Biol. 2018, 14, e1005911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borcherding, N.; Cole, K.; Kluz, P.; Jorgensen, M.; Kolb, R.; Bellizzi, A.M.; Zhang, W. Re-Evaluating E-Cadherin and β-Catenin. Am. J. Pathol. 2018, 188, 1910–1920. [Google Scholar] [CrossRef] [PubMed]
- Guilford, P.; Hopkins, J.; Harraway, J.; McLeod, M.; McLeod, N.; Harawira, P.; Taite, H.; Scoular, R.; Miller, A.; Reeve, A.E. E-cadherin germline mutations in familial gastric cancer. Nature 1998, 392, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Berx, G.; Van Roy, F. Involvement of Members of the Cadherin Superfamily in Cancer. Cold Spring Harb. Perspect. Biol. 2009, 1, a003129. [Google Scholar] [CrossRef]
- Fukagawa, A.; Ishii, H.; Miyazawa, K.; Saitoh, M. δEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells. Cancer Med. 2014, 4, 125–135. [Google Scholar] [CrossRef]
- Sánchez-Tilló, E.; Lázaro, A.; Torrent, R.; Cuatrecasas, M.; Vaquero, E.C.; Castells, A.; Engel, P.; Postigo, A. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 2010, 29, 3490–3500. [Google Scholar] [CrossRef] [Green Version]
- Grooteclaes, M.L.; Frisch, S.M. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 2000, 19, 3823–3828. [Google Scholar] [CrossRef] [Green Version]
- Wong, T.-S.; Gao, W.; Chan, J.Y.-W. Transcription Regulation of E-Cadherin by Zinc Finger E-Box Binding Homeobox Proteins in Solid Tumors. BioMed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Batlle, E.; Sancho, E.; Francí, C.; Domínguez, D.; Monfar, M.; Baulida, J.; De Herreros, A.G. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nature 2000, 2, 84–89. [Google Scholar] [CrossRef]
- Lin, Y.; Dong, C.; Zhou, B.P. Epigenetic regulation of EMT: The Snail story. Curr. Pharm. Des. 2014, 20, 1698–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosivatz, E.; Becker, I.; Specht, K.; Fricke, E.; Luber, B.; Busch, R.; Höfler, H.; Becker, K.-F. Differential Expression of the Epithelial-Mesenchymal Transition Regulators Snail, SIP1, and Twist in Gastric Cancer. Am. J. Pathol. 2002, 161, 1881–1891. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Huang, J.; Yang, W.; Lu, L.; Jin, H.; Wei, Z.; Yang, J.Y.; Arabnia, H.R.; Liu, J.S.; et al. The clinical significance of snail protein expression in gastric cancer: A meta-analysis. Hum. Genom. 2016, 10, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari-Amorotti, G.; Fragliasso, V.; Esteki, R.; Prudente, Z.; Soliera, A.R.; Cattelani, S.; Manzotti, G.; Grisendi, G.; Dominici, M.; Pieraccioli, M.; et al. Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res. 2012, 73, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Chen, H.-C.; Zhang, D.; Fu, J. Twist: A molecular target in cancer therapeutics. Tumor Biol. 2013, 34, 2497–2506. [Google Scholar] [CrossRef]
- Yang, F.; Sun, L.; Li, Q.; Han, X.; Lei, L.; Zhang, H.; Shang, Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2011, 31, 110–123. [Google Scholar] [CrossRef] [Green Version]
- Hanly, D.J.; Esteller, M.; Berdasco, M. Interplay between long non-coding RNAs and epigenetic machinery: Emerging targets in cancer? Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170074. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, B. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [Green Version]
- Shenas, M.H.M.; Eghbal-Fard, S.; Mehrisofiani, V.; Yazdani, N.A.; Farzam, O.R.; Marofi, F.; Yousefi, M. MicroRNAs and signaling networks involved in epithelial-mesenchymal transition. J. Cell. Physiol. 2018, 234, 5775–5785. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Sandoval-Bórquez, A.; Saavedra, K.; Carrasco-Avino, G.; García-Bloj, B.; Fry, J.; Wichmann, I.; Corvalán, A.H. Noncoding Genomics in Gastric Cancer and the Gastric Precancerous Cascade: Pathogenesis and Biomarkers. Dis. Markers 2015, 2015, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenzi, L.; Cobos, F.A.A.; Decock, A.; Everaert, C.; Helsmoortel, H.H.; Lefever, S.; Verboom, K.; Volders, P.-J.; Speleman, F.; Vandesompele, J.; et al. Long noncoding RNA expression profiling in cancer: Challenges and opportunities. Genes Chromosom. Cancer 2019, 58, 191–199. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudenas, B.L.; Wang, L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci. Rep. 2018, 8, 16385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhang, X.; Chen, W.; Hu, X.; Li, J.; Liu, C. Regulatory roles of long noncoding RNAs implicated in cancer hallmarks. Int. J. Cancer 2019, 146, 906–916. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2017, 18, 5–18. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Chow, J.T.-S.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Xue, M.; Du, S.; Feng, W.; Zhang, K.; Zhang, L.; Liu, H.; Jia, G.; Wu, L.; Hu, X.; et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat. Commun. 2019, 10, 1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, K.C.D.S.; Bona, A.B.; Da Silva, F.J.; Pinheiro, T.M.; Alcantara, D.D.F.A.; Lamarão, L.M.; Moreira-Nunes, C.A.; Assumpcao, P.P.; Burbano, R.M.R.; Calcagno, D.Q. Expression of hsa-miR-9 and MYC Copy Number Variation in Hereditary Diffuse Gastric Cancer. Anticancer Res. 2017, 37, 2401–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Önder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature 2010, 12, 247–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; He, L.; Li, T.; Lu, Y.; Miao, Y.; Liang, S.; Guo, H.; Bai, M.; Xie, H.; Luo, G.; et al. SRF expedites metastasis and modulates the epithelial to mesenchymal transition by regulating miR-199a-5p expression in human gastric cancer. Cell Death Differ. 2014, 21, 1900–1913. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Gao, Y.-Q. MiR-217 is involved in the carcinogenesis of gastric cancer by down-regulating CDH1 expression. Kaohsiung J. Med. Sci. 2018, 34, 377–384. [Google Scholar] [CrossRef]
- Yanaka, Y.; Muramatsu, T.; Uetake, H.; Kozaki, K.-I.; Inazawa, J. miR-544ainduces epithelial–mesenchymal transition through the activation of WNT signaling pathway in gastric cancer. Carcinogenesis 2015, 36, 1363–1371. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Wang, Y.; Shan, B.; Han, J.; Zhu, H.; Lv, Y.; Fan, X.; Sang, M.; Liu, X.-D.; Liu, W. The downregulation of miR-200c/141 promotes ZEB1/2 expression and gastric cancer progression. Med. Oncol. 2014, 32, 428. [Google Scholar] [CrossRef]
- Zhou, X.; Men, X.; Zhao, R.; Han, J.; Fan, Z.; Wang, Y.; Lv, Y.; Zuo, J.; Zhao, L.; Sang, M.; et al. miR-200c inhibits TGF-β-induced-EMT to restore trastuzumab sensitivity by targeting ZEB1 and ZEB2 in gastric cancer. Cancer Gene Ther. 2018, 25, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; He, X.; Xu, J.; Zhang, G.; Yang, Y.; Ma, J.; Sun, Y.; Ni, H.; Wang, F. Advantages of Restoring miR-205-3p Expression for Better Prognosis of Gastric Cancer via Prevention of Epithelial-mesenchymal Transition. J. Gastric Cancer 2020, 20, 212. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, R.; Zhang, S.; Xu, R.; Yang, Q. MicroRNA-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. Gene 2019, 700, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.-Q.; Jiang, S.-B.; He, X.-J.; Xia, Y.-J.; Hu, W.-J.; Luo, J.-G.; Zhang, J. MicroRNA-145-5p inhibits gastric cancer invasiveness through targeting N-cadherin and ZEB2 to suppress epithelial–mesenchymal transition. OncoTargets Ther. 2016, 9, 2305–2315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurashige, J.; Kamohara, H.; Watanabe, M.; Hiyoshi, Y.; Iwatsuki, M.; Tanaka, Y.; Kinoshita, K.; Saito, S.; Baba, Y.; Baba, H. MicroRNA-200b Regulates Cell Proliferation, Invasion, and Migration by Directly Targeting ZEB2 in Gastric Carcinoma. Ann. Surg. Oncol. 2012, 19, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Wu, Z.; Lin, L.; Zhou, M.; Wang, L.; Ma, H.; Xia, J.; Bin, J.; Liao, Y.; Liao, W. MiR-338-3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget 2015, 6, 15222–15234. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.-J.; Jiao, B.-P.; Liu, Y.-J.; Li, Y.-R.; Deng, B.-B. Reactivation of microRNA-506 inhibits gastric carcinoma cell metastasis through ZEB2. Aging 2019, 11, 1821–1831. [Google Scholar] [CrossRef]
- Zuo, Q.-F.; Cao, L.-Y.; Yu, T.; Gong, L.; Wang, L.-N.; Zhao, Y.-L.; Xiao, B.; Zou, Q. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015, 6, e2000. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, C. MiR-153 regulates metastases of gastric cancer through Snail. Tumor Biol. 2015, 37, 15509–15515. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Qiu, F.; Fu, M.; Chen, H.; Wang, H. Propofol Reduces Epithelial to Mesenchymal Transition, Invasion and Migration of Gastric Cancer Cells through the MicroRNA-195-5p/Snail Axis. Med. Sci. Monit. 2020, 26, e920981-1. [Google Scholar] [CrossRef]
- Liu, Z.; Long, J.; Du, R.; Ge, C.; Guo, K.; Xu, Y. miR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumor Biol. 2016, 37, 8327–8335. [Google Scholar] [CrossRef]
- Yu, T.; Wang, L.-N.; Li, W.; Zuo, Q.-F.; Li, M.-M.; Zou, Q.; Xiao, B. Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 2018, 109, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.-D.; Cheng, J.-T.; Chandoo, A.; Sun, X.-W.; Zhang, L.; Lu, M.-D.; Sun, W.-J.; Huang, Y.-P. microRNA-33a prevents epithelial-mesenchymal transition, invasion, and metastasis of gastric cancer cells through the Snail/Slug pathway. Am. J. Physiol. Liver Physiol. 2019, 317, G147–G160. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-L.; Gao, H.-L.; Lv, X.-K.; Hei, Y.-R.; Li, P.-Z.; Zhang, J.-X.; Lu, N. MicroRNA-124 inhibits cell invasion and epithelial-mesenchymal transition by directly repressing Snail2 in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3389–3396. [Google Scholar] [PubMed]
- Yang, L.; Liang, H.; Wang, Y.; Gao, S.; Yin, K.; Liu, Z.; Zheng, X.; Lv, Y.; Wang, L.; Zhang, C.-Y.; et al. MiRNA-203 suppresses tumor cell proliferation, migration and invasion by targeting Slug in gastric cancer. Protein Cell 2016, 7, 383–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakimura, S.; Sugimachi, K.; Kurashige, J.; Ueda, M.; Hirata, H.; Nambara, S.; Komatsu, H.; Saito, T.; Takano, Y.; Uchi, R.; et al. The miR-506-Induced Epithelial–Mesenchymal Transition is Involved in Poor Prognosis for Patients with Gastric Cancer. Ann. Surg. Oncol. 2015, 22, 1436–1443. [Google Scholar] [CrossRef]
- Wang, T.; Hou, J.; Li, Z.; Zheng, Z.; Wei, J.; Song, D.; Hu, T.; Wu, Q.; Yang, J.Y.; Cai, J.-C. miR-15a-3p and miR-16-1-3p Negatively Regulate Twist1 to Repress Gastric Cancer Cell Invasion and Metastasis. Int. J. Biol. Sci. 2017, 13, 122–134. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.; Sun, D.; Zhang, L.; Song, L. miR-186 affects the proliferation, invasion and migration of human gastric cancer by inhibition of Twist1. Oncotarget 2016, 7, 79956–79963. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Li, X.; Guo, Z.; Zhou, F. MicroRNA-381 regulates the growth of gastric cancer cell by targeting TWIST1. Mol. Med. Rep. 2019, 20, 4376–4382. [Google Scholar] [CrossRef]
- Liu, C.; Jian, M.; Qi, H.; Mao, W.-Z. MicroRNA 495 Inhibits Proliferation and Metastasis and Promotes Apoptosis by Targeting Twist1 in Gastric Cancer Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 27, 389–397. [Google Scholar] [CrossRef]
- Qi, F.; Liu, X.; Wu, H.; Yu, X.; Wei, C.; Huang, X.; Ji, G.; Nie, F.; Wang, K. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J. Hematol. Oncol. 2017, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Huang, F.; Wang, H.; Cheng, F.; Pi, Y.; Zhao, J.; Li, Z. Knockdown of TPT1-AS1 inhibits cell proliferation, cell cycle G1/S transition, and epithelial-mesenchymal transition in gastric cancer. Bosn. J. Basic Med. Sci. 2020. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Liu, H.-M.; Wu, W.-H.; Liu, H.; Pan, Y.; Li, W.-J. Upregulation of long noncoding RNA CCAT1-L promotes epithelial–mesenchymal transition in gastric adenocarcinoma. OncoTargets Ther. 2018, 11, 5647–5655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liu, L.; Wan, J.-X.; Song, Y. Long noncoding RNA SNHG20 promotes gastric cancer progression by inhibiting p21 expression and regulating the GSK-3β/ β-catenin signaling pathway. Oncotarget 2017, 8, 80700–80708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Huang, L.; Lu, X.; Wang, K.; Ning, X.; Liu, Z. Upregulated expression of MNX1-AS1 long noncoding RNA predicts poor prognosis in gastric cancer. Bosn. J. Basic Med. Sci. 2019, 19, 164–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.-W.; Kong, Y.; Sun, X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 1571–1579. [Google Scholar] [CrossRef]
- Li, Z.; Ding, Z.; Rong, D.; Tang, W.; Cao, H. Overexpression of lncRNA AFAP1-AS1 promotes cell proliferation and invasion in gastric cancer. Oncol. Lett. 2019, 18, 3211–3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Wang, Z.-G.; Chi, T.-S. Long noncoding RNA Lnc01614 promotes the occurrence and development of gastric cancer by activating EMT pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1307–1314. [Google Scholar]
- Zhang, K.; Lu, C.; Huang, X.; Cui, J.; Li, J.; Gao, Y.; Liang, W.; Liu, Y.; Sun, Y.; Liu, H.; et al. Long noncoding RNA AOC4P regulates tumor cell proliferation and invasion by epithelial–mesenchymal transition in gastric cancer. Ther. Adv. Gastroenterol. 2019, 12, 1756284819827697. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Wu, N.; Zhang, Z.; Zhong, X.; Zhang, H.; Guo, H.; Nie, Y.; Liu, Y. Long Non-coding RNA LINC00941 as a Potential Biomarker Promotes the Proliferation and Metastasis of Gastric Cancer. Front. Genet. 2019, 10, 5. [Google Scholar] [CrossRef] [Green Version]
- Zhang, E.; He, X.; Yin, D.; Han, L.; Qiu, M.; Xu, T.; Xia, R.; Xu, L.; Yin, R.; De, W. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016, 7, e2109. [Google Scholar] [CrossRef]
- Jiao, J.; Zhang, S. Long non-coding RNA MEG-3 suppresses gastric carcinoma cell growth, invasion and migration via EMT regulation. Mol. Med. Rep. 2019, 20, 2685–2693. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.; Si, S.; Zhang, Q.; Li, C.; Zhao, F.; Wang, F.; Yu, J.; Ma, R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J. Exp. Clin. Cancer Res. 2015, 34, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Wu, J.; Huang, W.; Peng, J.; Ye, J.; Yang, L.; Yuan, Y.; Chen, C.; Zhang, C.; Cai, S.; et al. Long Non-Coding RNA RP11-789C1.1 Suppresses Epithelial to Mesenchymal Transition in Gastric Cancer through the RP11-789C1.1/MiR-5003/E-Cadherin Axis. Cell. Physiol. Biochem. 2018, 47, 2432–2444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, K.; Wang, T.; Cui, J.; Xi, H.; Wang, Y.; Song, Y.; Zhao, X.; Wei, B.; Chen, L. Long non-coding RNA AFAP1-antisense RNA 1 promotes the proliferation, migration and invasion of gastric cancer cells and is associated with poor patient survival. Oncol. Lett. 2018, 15, 8620–8626. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, C.-S.; Li, S.-J.; Li, Z.; Sun, F.-B. LncRNA LOC554202 promotes proliferation and migration of gastric cancer cells through regulating p21 and E-cadherin. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8690–8697. [Google Scholar]
- Wang, S.; Ni, B.; Zhang, Z.; Wang, C.; Wo, L.; Zhou, C.; Zhao, Q.; Zhao, E. Long non-coding RNA DNM3OS promotes tumor progression and EMT in gastric cancer by associating with Snail. Biochem. Biophys. Res. Commun. 2019, 511, 57–62. [Google Scholar] [CrossRef]
- Xu, Z.-Y.; Yu, Q.-M.; Du, Y.-A.; Yang, L.-T.; Dong, R.-Z.; Huang, L.; Yu, P.-F.; Cheng, X.-D. Knockdown of Long Non-coding RNA HOTAIR Suppresses Tumor Invasion and Reverses Epithelial-mesenchymal Transition in Gastric Cancer. Int. J. Biol. Sci. 2013, 9, 587–597. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.K.; Lee, J.H.; Ivan, C.; Ling, H.; Zhang, X.; Park, C.H.; Calin, G.A.; Kil Lee, S. MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer 2017, 17, 46. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Y.; Chen, Z.-H.; Peng, J.-J.; Wu, J.-L.; Yuan, Y.; Zhai, E.-T.; Cai, S.-R.; He, Y.-L.; Song, W. Up-regulation of long non-coding RNA XLOC_010235 regulates epithelial-to-mesenchymal transition to promote metastasis by associating with Snail1 in gastric cancer. Sci. Rep. 2017, 7, 2461. [Google Scholar] [CrossRef] [Green Version]
- Fu, M.; Huang, Z.; Zang, X.; Pan, L.; Liang, W.; Chen, J.; Qian, H.; Xu, W.; Jiang, P.; Zhang, X. Long noncoding RNA LINC00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Prolif. 2017, 51, e12425. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Shen, Z.; Wang, B.; Ye, C.; Lai, Z.; Jiang, H.; Wang, Z.; Jiang, K.; Ye, Y.; Wang, S. Long non-coding RNA GPR65-1 is up-regulated in gastric cancer and promotes tumor growth through the PTEN-AKT-slug signaling pathway. Cell Cycle 2018, 17, 759–765. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Chen, J.; He, B.; Li, Q.; Li, Y.-D.; Gao, Y. A FOXM1 related long non-coding RNA contributes to gastric cancer cell migration. Mol. Cell. Biochem. 2015, 406, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, F.; Chen, H.; Tan, Q.; Qiu, S.; Chen, S.; Jing, W.; Yu, M.; Liang, C.; Ye, S.; et al. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging 2016, 8, 2023–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Li, L.; Zheng, Z.; Chen, S.; Chen, E.; Hu, Y. Long non-coding RNA linc00261 suppresses gastric cancer progressionviapromoting Slug degradation. J. Cell. Mol. Med. 2016, 21, 955–967. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Xiang, S.; Ma, J.; Hui, P.; Wang, T.; Meng, W.; Shi, M.; Wang, Y. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol. Oncol. 2018, 12, 799–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, G.; Quan, J.; Dong, D.; Wang, Q. Long Noncoding RNA CAT104 Promotes Cell Viability, Migration, and Invasion in Gastric Carcinoma Cells Through Activation of MicroRNA-381-Inhibiting Zinc Finger E-box-Binding Homeobox 1 (ZEB1) Expression. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ye, F.; Yin, C.; Zhuang, Y.; Yue, G.; Zhang, G. The Interaction Between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cell. Physiol. Biochem. 2015, 36, 1440–1452. [Google Scholar] [CrossRef]
- Li, D.; Wang, J.; Zhang, M.; Hu, X.; She, J.; Qiu, X.; Zhang, X.; Xu, L.; Liu, Y.; Qin, S. LncRNA MAGI2-AS3 Is Regulated by BRD4 and Promotes Gastric Cancer Progression via Maintaining ZEB1 Overexpression by Sponging miR-141/200a. Mol. Ther. Nucleic Acids 2020, 19, 109–123. [Google Scholar] [CrossRef]
- Yan, K.; Tian, J.; Shi, W.; Xia, H.; Zhu, Y. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell. Physiol. Biochem. 2017, 42, 999–1012. [Google Scholar] [CrossRef]
- Ma, M.-H.; An, J.-X.; Zhang, C.; Liu, J.; Liang, Y.; Zhang, C.-D.; Zhang, Z.; Dai, D.-Q. ZEB1-AS1 initiates a miRNA-mediated ceRNA network to facilitate gastric cancer progression. Cancer Cell Int. 2019, 19, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Gong, P.; Qiao, F.; Wu, H.; Cui, H.; Li, Y.; Zheng, Y.; Zhou, M.; Fan, H. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018, 9, 1158. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, R.; Sun, Y.; Liu, H.; Zhang, H.; Sun, Y.; Liu, L.; Li, Y.; Song, L.; Gao, P. SP1-activated long noncoding RNA lncRNA GCMA functions as a competing endogenous RNA to promote tumor metastasis by sponging miR-124 and miR-34a in gastric cancer. Oncogene 2020, 39, 4854–4868. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Lv, L.; Liao, S. Long non-coding RNA H19 regulates cell growth and metastasis via the miR-22-3p/Snail1 axis in gastric cancer. Int. J. Oncol. 2019, 54, 2157–2168. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xiao, B.; Yu, T.; Gong, L.; Wang, Y.; Zhang, X.; Zou, Q.; Zuo, Q. lncRNA PVT1 promotes the migration of gastric cancer by functioning as ceRNA of miR-30a and regulating Snail. J. Cell. Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yuan, Y.; Zhang, Y.; Cheng, L.; Zhou, X.; Chen, K. SNHG7 accelerates cell migration and invasion through regulating miR-34a-Snail-EMT axis in gastric cancer. Cell Cycle 2019, 19, 142–152. [Google Scholar] [CrossRef]
- Chen, D.-L.; Ju, H.-Q.; Lu, Y.-X.; Chen, L.-Z.; Zeng, Z.-L.; Zhang, D.-S.; Luo, H.-Y.; Wang, F.; Qiu, M.-Z.; Wang, D.-S.; et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J. Exp. Clin. Cancer Res. 2016, 35, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Lei, K.; Liang, X.; Gao, Y.; Xu, B.; Xu, Y.; Li, Y.; Tao, Y.; Shi, W.; Liu, J.-W. Lnc-ATB contributes to gastric cancer growth through a MiR-141-3p/TGFβ2 feedback loop. Biochem. Biophys. Res. Commun. 2017, 484, 514–521. [Google Scholar] [CrossRef]
- Song, F.; Yang, D.; Liu, B.; Guo, Y.; Zheng, H.; Li, L.; Wang, T.; Yu, J.; Zhao, Y.; Niu, R.; et al. Integrated MicroRNA Network Analyses Identify a Poor-Prognosis Subtype of Gastric Cancer Characterized by the miR-200 Family. Clin. Cancer Res. 2013, 20, 878–889. [Google Scholar] [CrossRef] [Green Version]
- Riquelme, I.; Letelier, P.; Riffo-Campos, A.L.; Brebi, P.; Roa, J.C. Emerging Role of miRNAs in the Drug Resistance of Gastric Cancer. Int. J. Mol. Sci. 2016, 17, 424. [Google Scholar] [CrossRef] [Green Version]
- Saito, T.; Kurashige, J.; Nambara, S.; Komatsu, H.; Hirata, H.; Ueda, M.; Sakimura, S.; Uchi, R.; Takano, Y.; Shinden, Y.; et al. A Long Non-coding RNA Activated by Transforming Growth Factor-β is an Independent Prognostic Marker of Gastric Cancer. Ann. Surg. Oncol. 2015, 22, 915–922. [Google Scholar] [CrossRef]
- Imamura, T.; Komatsu, S.; Ichikawa, D.; Miyamae, M.; Okajima, W.; Ohashi, T.; Kiuchi, J.; Nishibeppu, K.; Kosuga, T.; Konishi, H.; et al. Low plasma levels of miR-101 are associated with tumor progression in gastric cancer. Oncotarget 2017, 8, 106538–106550. [Google Scholar] [CrossRef] [Green Version]
- Cata, J.P.; Owusu-Agyemang, P.; Kapoor, R.; Lonnqvist, P.-A. Impact of Anesthetics, Analgesics, and Perioperative Blood Transfusion in Pediatric Cancer Patients. Anesth. Analg. 2019, 129, 1653–1665. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Gao, J.; Yan, N.; Wu, B.; Ren, Y.; Li, H.; Liang, J. Propofol inhibits the growth and survival of gastric cancer cells in vitro through the upregulation of ING3. Oncol. Rep. 2016, 37, 587–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Liu, Y.; Huang, L.; Zhang, F.; Kang, R. Effects of propofol on cancer development and chemotherapy: Potential mechanisms. Eur. J. Pharmacol. 2018, 831, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-B.; Bantounas, I.; Lee, D.-Y.; Phylactou, L.; A Caldwell, M.; Uney, J.B. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2008, 37, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Mitra, R.; Chen, X.; Greenawalt, E.J.; Maulik, U.; Jiang, W.; Zhao, Z.; Eischen, C.M. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat. Commun. 2017, 8, 1604. [Google Scholar] [CrossRef] [Green Version]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [Green Version]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef] [Green Version]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2015, 44, 863–877. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.-Q.; Li, X.-Y.; Hu, C.Y.; Ford, M.; Kleer, C.G.; Weiss, S.J. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc. Natl. Acad. Sci. USA 2012, 109, 16654–16659. [Google Scholar] [CrossRef] [Green Version]
- Hansji, H.; Leung, E.; Baguley, B.C.; Finlay, G.; Cameron-Smith, D.; Figueiredo, V.C.; Askarian-Amiri, M. ZFAS1: A long noncoding RNA associated with ribosomes in breast cancer cells. Biol. Direct 2016, 11, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Liang, W.; Fu, M.; Huang, Z.-H.; Li, X.; Zhang, X.; Zhang, P.; Qian, H.; Jiang, P.-C.; Xu, W.; et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 2017, 17, 878–1004. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk—Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landeros, N.; Santoro, P.M.; Carrasco-Avino, G.; Corvalan, A.H. Competing Endogenous RNA Networks in the Epithelial to Mesenchymal Transition in Diffuse-Type of Gastric Cancer. Cancers 2020, 12, 2741. https://doi.org/10.3390/cancers12102741
Landeros N, Santoro PM, Carrasco-Avino G, Corvalan AH. Competing Endogenous RNA Networks in the Epithelial to Mesenchymal Transition in Diffuse-Type of Gastric Cancer. Cancers. 2020; 12(10):2741. https://doi.org/10.3390/cancers12102741
Chicago/Turabian StyleLanderos, Natalia, Pablo M. Santoro, Gonzalo Carrasco-Avino, and Alejandro H. Corvalan. 2020. "Competing Endogenous RNA Networks in the Epithelial to Mesenchymal Transition in Diffuse-Type of Gastric Cancer" Cancers 12, no. 10: 2741. https://doi.org/10.3390/cancers12102741
APA StyleLanderos, N., Santoro, P. M., Carrasco-Avino, G., & Corvalan, A. H. (2020). Competing Endogenous RNA Networks in the Epithelial to Mesenchymal Transition in Diffuse-Type of Gastric Cancer. Cancers, 12(10), 2741. https://doi.org/10.3390/cancers12102741