Microenvironmental Hypoxia Induces Dynamic Changes in Lung Cancer Synthesis and Secretion of Extracellular Vesicles
Abstract
:Simple Summary
Abstract
1. Introduction
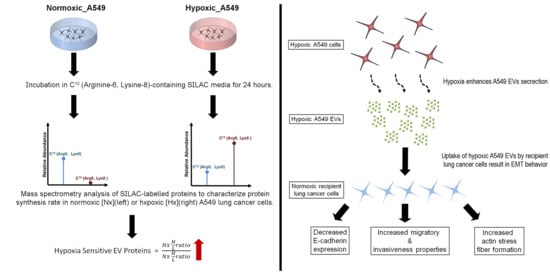
2. Materials and Methods
2.1. Cell Culture
2.2. Heavy Isotope Pulse/Trace for Hypoxia-Sensitive EV Proteins
2.3. Physical Characterisation of EVs
2.4. Proteomic Profiling of A549 EVs and Cell Lysate by pSILAC
2.5. A549 EVs Treatment on Lung Cancer Cells
2.6. Western Blots
2.7. Total RNA Extraction and Real-Time Quantitative PCR
2.8. Fluorescence Microscopy
2.9. Wound Healing and Migration Assay
2.10. Invasion Assay
2.11. Transmission Electron Microscopy
2.12. Statistical Analysis
3. Results
3.1. Oxygen Deprivation Promotes EV Secretion by A549 Human Lung Cancer Cells
3.2. Hypoxia Stress in Lung Cancer Cells Induces De Novo Synthesis of Specific EV Proteins
3.3. Altered Dynamics of Cancer EV Production and Release during Hypoxia Stress
3.4. Lung Cancer-Derived EVs Promote Epithelial-Mesenchymal Transition (EMT)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Semenza, G.L. Hypoxia, clonal selection, and the role of HIF-1 in tumor progression. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 71–103. [Google Scholar] [CrossRef]
- Klein, T.J.; Glazer, P.M. The tumor microenvironment and DNA repair. Semin. Radiat. Oncol. 2010, 20, 282–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics 2010, 9, 1085–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denzer, K.; Kleijmeer, M.J.; Heijnen, H.F.; Stoorvogel, W.; Geuze, H.J. Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 2000, 113, 3365–3374. [Google Scholar] [PubMed]
- Vella, L.J. The emerging role of exosomes in epithelial-mesenchymal-transition in cancer. Front. Oncol. 2014, 4, 361. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.G.; Grizzle, W.E. Exosomes and cancer: A newly described pathway of immune suppression. Clin. Cancer Res. 2011, 17, 959–964. [Google Scholar] [CrossRef] [Green Version]
- Park, J.E.; Dutta, B.; Tse, S.W.; Gupta, N.; Tan, C.F.; Low, J.K.; Yeoh, K.W.; Kon, O.L.; Tam, J.P.; Sze, S.K. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 2019, 38, 5158–5173. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Barry, S.; Kmetz, D.; Egger, M.; Pan, J.; Rai, S.N.; Qu, J.; McMasters, K.M.; Hao, H. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016, 376, 318–327. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef]
- Savagner, P.; Yamada, K.M.; Thiery, J.P. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J. Cell Bio. 1997, 137, 1403–1419. [Google Scholar] [CrossRef]
- Ogunwobi, O.O.; Liu, C. Hepatocyte growth factor upregulation promotes carcinogenesis and epithelial-mesenchymal transition in hepatocellular carcinoma via Akt and COX-2 pathways. Clin. Exp. Metastasis 2011, 28, 721–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vincan, E.; Barker, N. The upstream components of the Wnt signalling pathway in the dynamic EMT and MET associated with colorectal cancer progression. Clin. Exp. Metastasis 2008, 25, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Kong, D.; Sarkar, F.H. The role of Notch signaling pathway in epithelial-mesenchymal transition (EMT) during development and tumor aggressiveness. Curr. Drug Targets 2010, 11, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, C.H. Signaling networks guiding epithelial-mesenchymal transitions during embryogenesis and cancer progression. Cancer Sci. 2007, 98, 1512–1520. [Google Scholar] [CrossRef]
- Tan, C.F.; Teo, H.S.; Park, J.E.; Dutta, B.; Tse, S.W.; Leow, M.K.; Wahli, W.; Sze, S.K. Exploring Extracellular Vesicles Biogenesis in Hypothalamic Cells through a Heavy Isotope Pulse/Trace Proteomic Approach. Cells 2020, 9, 1320. [Google Scholar] [CrossRef]
- Bobadilla, A.V.P.; Arevalo, J.; Sarro, E.; Byrne, H.M.; Maini, P.K.; Carraro, T.; Balocco, S.; Meseguer, A.; Alarcon, T. In vitro cell migration quantification method for scratch assays. J. R. Soc. Interface 2019, 16, 20180709. [Google Scholar] [CrossRef] [Green Version]
- Oono, K.; Takahashi, K.; Sukehara, S.; Kurosawa, H.; Matsumura, T.; Taniguchi, S.; Ohta, S. Inhibition of PC3 human prostate cancer cell proliferation, invasion and migration by eicosapentaenoic acid and docosahexaenoic acid. Mol. Clin. Oncol. 2017, 7, 217–220. [Google Scholar] [CrossRef] [Green Version]
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ge, C.; Zhao, F.; Yan, M.; Hu, C.; Jia, D.; Tian, H.; Zhu, M.; Chen, T.; Jiang, G.; et al. Hypoxia-inducible factor 1 alpha-activated angiopoietin-like protein 4 contributes to tumor metastasis via vascular cell adhesion molecule-1/integrin β1 signaling in human hepatocellular carcinoma. Hepatology 2011, 54, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Fuady, J.H.; Bordoli, M.R.; Abreu-Rodríguez, I.; Kristiansen, G.; Hoogewijs, D.; Stiehl, D.P.; Wenger, R.H. Hypoxia-inducible factor-mediated induction of WISP-2 contributes to attenuated progression of breast cancer. Hypoxia (Auckl) 2014, 2, 23–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruf, M.; Mittmann, C.; Nowicka, A.M.; Hartmann, A.; Hermanns, T.; Poyet, C.; van den Broek, M.; Sulser, T.; Moch, H.; Schraml, P. pVHL/HIF-regulated CD70 expression is associated with infiltration of CD27+ lymphocytes and increased serum levels of soluble CD27 in clear cell renal cell carcinoma. Clin. Cancer Res. 2015, 21, 889–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, Y.; Zemans, R.; Correll, K.; Yang, I.V.; Ahmad, A.; Gao, B.; Mason, R.J. Stanniocalcin-1 is induced by hypoxia inducible factor in rat alveolar epithelial cells. Biochem. Biophys Res. Commun. 2014, 452, 1091–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, G.; Rangaswami, H.; Jain, S.; Kundu, G.C. Hypoxia regulates cross-talk between Syk and Lck leading to breast cancer progression and angiogenesis. J. Biol. Chem. 2006, 281, 11322–11331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolfs, A.; Kvietikova, I.; Gassmann, M.; Wenger, R.H. Oxygen-regulated transferrin expression is mediated by hypoxia-inducible factor-1. J. Biol. Chem. 1997, 272, 20055–20062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, N.V.; Kotch, L.E.; Agani, F.; Leung, S.W.; Laughner, E.; Wenger, R.H.; Gassmann, M.; Gearhart, J.D.; Lawler, A.M.; Yu, A.Y.; et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998, 12, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Feldser, D.; Agani, F.; Iyer, N.V.; Pak, B.; Ferreira, G.; Semenza, G.L. Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer Res. 1999, 59, 3915–3918. [Google Scholar]
- Pennacchietti, S.; Michieli, P.; Galluzzo, M.; Mazzone, M.; Giordano, S.; Comoglio, P.M. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 2003, 3, 347–361. [Google Scholar] [CrossRef] [Green Version]
- Gilkes, D.M.; Bajpai, S.; Wong, C.C.; Chaturvedi, P.; Hubbi, M.E.; Wirtz, D.; Semenza, G.L. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol. Cancer Res. 2013, 11, 456–466. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Jamindar, T.M.; Dawson, G. Hypoxia alters iron homeostasis and induces ferritin synthesis in oligodendrocytes. J. Neurochem. 1995, 64, 2458–2464. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Connolly, E.; Braunstein, S.; Formenti, S.; Schneider, R.J. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell Biol. 2006, 26, 3955–3965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fares, J.; Kashyap, R.; Zimmermann, P. Syntenin: Key player in cancer exosome biogenesis and uptake? Cell Adh. Migr. 2017, 11, 124–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavallaro, U.; Christofori, G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer 2004, 4, 118–132. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, G.; Li, X.; Zhang, Y.; Jiang, Y.; Shen, J.; Liu, J.; Wang, Q.; Zhu, J.; Feng, X.; et al. Hypoxia induces epithelial-mesenchymal transition via activation of SNAI1 by hypoxia-inducible factor -1alpha in hepatocellular carcinoma. BMC Cancer 2013, 13, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haynes, J.; Srivastava, J.; Madson, N.; Wittmann, T.; Barber, D.L. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol. Biol. Cell 2011, 22, 4750–4764. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Mandard, S.; Zandbergen, F.; van Straten, E.; Wahli, W.; Kuipers, F.; Muller, M.; Kersten, S. The fasting-induced adipose factor/angiopoietin-like protein 4 is physically associated with lipoproteins and governs plasma lipid levels and adiposity. J. Biol. Chem. 2006, 281, 934–944. [Google Scholar] [CrossRef] [Green Version]
- Ma, T.; Jham, B.C.; Hu, J.; Friedman, E.R.; Basile, J.R.; Molinolo, A.; Sodhi, A.; Montaner, S. Viral G protein-coupled receptor up-regulates Angiopoietin-like 4 promoting angiogenesis and vascular permeability in Kaposi’s sarcoma. Proc. Natl. Acad. Sci. USA 2010, 107, 14363–14368. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Oike, Y.; Yasunaga, K.; Hamada, K.; Miyata, K.; Matsumoto, S.; Sugano, S.; Tanihara, H.; Masuho, Y.; Suda, T. Inhibition of angiogenesis and vascular leakiness by angiopoietin-related protein 4. Cancer Res. 2003, 63, 6651–6657. [Google Scholar] [PubMed]
- Babapoor-Farrokhran, S.; Jee, K.; Puchner, B.; Hassan, S.J.; Xin, X.; Rodrigues, M.; Kashiwabuchi, F.; Ma, T.; Hu, K.; Deshpande, M.; et al. Angiopoietin-like 4 is a potent angiogenic factor and a novel therapeutic target for patients with proliferative diabetic retinopathy. Proc. Natl. Acad Sci. USA 2015, 112, E3030–E3039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.Y.; Kuang, J.L.; Yan, C.S.; Tu, X.Y.; Zhao, J.H.; Cheng, X.S.; Ye, X.Q. NRSN2 promotes non-small cell lung cancer cell growth through PI3K/Akt/mTOR pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 2574–2581. [Google Scholar] [PubMed]
- Banerjee, S.; Dhar, G.; Haque, I.; Kambhampati, S.; Mehta, S.; Sengupta, K.; Tawfik, O.; Phillips, T.A.; Banerjee, S.K. CCN5/WISP-2 expression in breast adenocarcinoma is associated with less frequent progression of the disease and suppresses the invasive phenotypes of tumor cells. Cancer Res. 2008, 68, 7606–7612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritah, A.; Saucier, C.; De Wever, O.; Bracke, M.; Bieche, I.; Lidereau, R.; Gespach, C.; Drouot, S.; Redeuilh, G.; Sabbah, M. Role of WISP-2/CCN5 in the maintenance of a differentiated and noninvasive phenotype in human breast cancer cells. Mol. Cell Biol. 2008, 28, 1114–1123. [Google Scholar] [CrossRef] [Green Version]
- Tambe, Y.; Hasebe, M.; Kim, C.J.; Yamamoto, A.; Inoue, H. The drs tumor suppressor regulates glucose metabolism via lactate dehydrogenase-B. Mol. Carcinog. 2016, 55, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Krystal, G.W.; DeBerry, C.S.; Linnekin, D.; Litz, J. Lck associates with and is activated by Kit in a small cell lung cancer cell line: Inhibition of SCF-mediated growth by the Src family kinase inhibitor PP1. Cancer Res. 1998, 58, 4660–4666. [Google Scholar]
- Jin, D.; Tao, J.; Li, D.; Wang, Y.; Li, L.; Hu, Z.; Zhou, Z.; Chang, X.; Qu, C.; Zhang, H. Golgi protein 73 activation of MMP-13 promotes hepatocellular carcinoma cell invasion. Oncotarget 2015, 6, 33523–33533. [Google Scholar] [CrossRef] [Green Version]
- Li, R.M.; Nai, M.M.; Duan, S.J.; Li, S.X.; Yin, B.N.; An, F.; Zhai, Y.Q.; Liu, J.; Chu, Y.R.; Yu, Y.; et al. Down-expression of GOLM1 enhances the chemo-sensitivity of cervical cancer to methotrexate through modulation of the MMP13/EMT axis. Am. J. Cancer Res. 2018, 8, 964–980. [Google Scholar]
- Yang, Y.; Liu, Q.; Zhang, H.; Zhao, H.; Mao, R.; Li, Z.; Ya, S.; Jia, C.; Bao, Y. Silencing of GP73 inhibits invasion and metastasis via suppression of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncol. Rep. 2017, 37, 1182–1188. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.H.; Zhu, W.W.; Zhang, J.B.; Qin, Y.; Lu, M.; Lin, G.L.; Guo, L.; Zhang, B.; Lin, Z.H.; Roessler, S.; et al. GOLM1 Modulates EGFR/RTK Cell-Surface Recycling to Drive Hepatocellular Carcinoma Metastasis. Cancer Cell 2016, 30, 444–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015, 126, 11–51. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zheng, S.; Wang, J.; Long, H.; Fang, L.; Wang, G.; Li, Z.; Que, T.; Liu, Y.; Li, Y.; et al. Hypoxia-induced PLOD2 promotes proliferation, migration and invasion via PI3K/Akt signaling in glioma. Oncotarget 2017, 8, 41947–41962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Zhang, L.; Wei, Y.; Zhang, X.; Xu, R.; Han, M.; Huang, B.; Chen, A.; Li, W.; Zhang, Q.; et al. Procollagen-lysine 2-oxoglutarate 5-dioxygenase 2 promotes hypoxia-induced glioma migration and invasion. Oncotarget 2017, 8, 23401–23413. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, J.; Hu, G.; Liu, L.; Liang, W. Hypoxia and TGF-beta1 induced PLOD2 expression improve the migration and invasion of cervical cancer cells by promoting epithelial-to-mesenchymal transition (EMT) and focal adhesion formation. Cancer Cell Int. 2017, 17, 54. [Google Scholar] [CrossRef]
- Du, H.; Chen, Y.; Hou, X.; Huang, Y.; Wei, X.; Yu, X.; Feng, S.; Wu, Y.; Zhan, M.; Shi, X.; et al. PLOD2 regulated by transcription factor FOXA1 promotes metastasis in NSCLC. Cell Death Dis. 2017, 8, e3143. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, H.; Terajima, M.; Banerjee, P.; Liu, X.; Yu, J.; Momin, A.A.; Katayama, H.; Hanash, S.M.; Burns, A.R.; et al. Lysyl Hydroxylase 2 Is Secreted by Tumor Cells and Can Modify Collagen in the Extracellular Space. J. Biol. Chem. 2016, 291, 25799–25808. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Gu, L.; Li, H.; Gao, Y.; Li, X.; Shen, D.; Gong, H.; Li, S.; Niu, S.; Zhang, Y.; et al. Hypoxia-induced overexpression of stanniocalcin-1 is associated with the metastasis of early stage clear cell renal cell carcinoma. J. Transl. Med. 2015, 13, 56. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.K.; Leung, C.O.; Wong, C.C.; Ho, D.W.; Chok, K.S.; Lai, C.L.; Ng, I.O.; Lo, R.C. Secretory Stanniocalcin 1 promotes metastasis of hepatocellular carcinoma through activation of JNK signaling pathway. Cancer Lett. 2017, 403, 330–338. [Google Scholar] [CrossRef]
- Li, Y.; He, Z.C.; Zhang, X.N.; Liu, Q.; Chen, C.; Zhu, Z.; Chen, Q.; Shi, Y.; Yao, X.H.; Cui, Y.H.; et al. Stanniocalcin-1 augments stem-like traits of glioblastoma cells through binding and activating NOTCH1. Cancer Lett. 2018, 416, 66–74. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; Wu, Y.; Wu, J.; Pang, S.; Pan, R.; Wen, T. A novel function of dcf1 during the differentiation of neural stem cells in vitro. Cell Mol. NeuroBiol. 2008, 28, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.; Deschoolmeester, V.; Zwaenepoel, K.; Rolfo, C.; Silence, K.; Rottey, S.; Lardon, F.; Smits, E.; Pauwels, P. CD70: An emerging target in cancer immunotherapy. Pharmacol. Ther. 2015, 155, 1–10. [Google Scholar] [CrossRef] [PubMed]
- DeBarros, A.; Chaves-Ferreira, M.; d’Orey, F.; Ribot, J.C.; Silva-Santos, B. CD70-CD27 interactions provide survival and proliferative signals that regulate T cell receptor-driven activation of human gammadelta peripheral blood lymphocytes. Eur. J. Immunol. 2011, 41, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Pich, C.; Sarrabayrouse, G.; Teiti, I.; Mariame, B.; Rochaix, P.; Lamant, L.; Favre, G.; Maisongrosse, V.; Tilkin-Mariame, A.F. Melanoma-expressed CD70 is involved in invasion and metastasis. Br. J. Cancer 2016, 114, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Kucharzewska, P.; Belting, M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Yu, D.D.; Wu, Y.; Shen, H.Y.; Lv, M.M.; Chen, W.X.; Zhang, X.H.; Zhong, S.L.; Tang, J.H.; Zhao, J.H. Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 2015, 106, 959–964. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Wheelock, M.J.; Shintani, Y.; Maeda, M.; Fukumoto, Y.; Johnson, K.R. Cadherin switching. J. Cell Sci. 2008, 121, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Gulhati, P.; Bowen, K.A.; Liu, J.; Stevens, P.D.; Rychahou, P.G.; Chen, M.; Lee, E.Y.; Weiss, H.L.; O’Connor, K.L.; Gao, T.; et al. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011, 71, 3246–3256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunaga, N.; Miyajima, K.; Suzuki, M.; Sato, M.; White, M.A.; Ramirez, R.D.; Shay, J.W.; Gazdar, A.F.; Minna, J.D. Different roles for caveolin-1 in the development of non-small cell lung cancer versus small cell lung cancer. Cancer Res. 2004, 64, 4277–4285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, E.K.; Wong, B.Y.; Yau, T.O.; Ng, I.O. Upregulation of the Wnt co-receptor LRP6 promotes hepatocarcinogenesis and enhances cell invasion. PLoS ONE 2012, 7, e36565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, Y.; Zhang, J.; Zhong, J.; Yang, R. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol. Med. Rep. 2018, 17, 5037–5042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kim, T.Y.; Lee, M.S.; Mun, J.Y.; Ihm, C.; Kim, S.A. Exosome cargo reflects TGF-beta1-mediated epithelial-to-mesenchymal transition (EMT) status in A549 human lung adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2016, 478, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [Green Version]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [Green Version]





| Protein Names (Gene Symbols) | A549 Cells (Nx) | A549 EVs (Nx) | A549 Cells (Hx) | A549 EVs (Hx) | Hypoxia Inducible Gene | ||||
|---|---|---|---|---|---|---|---|---|---|
| H/L | emPAI | H/L | emPAI | H/L | emPAI | H/L | emPAI | ||
| Angiopoietin-Related Protein 4 (ANGPTL4) | - | - | - | - | - | 0.122 | 100 | 0.413 | Yes [21] |
| Neurensin 2 (NRSN2) | - | - | - | - | - | - | 100 | 0.233 | - |
| WNT1-Inducible-Signaling Pathway Protein 2 (WISP2) | - | - | - | - | - | - | 100 | 0.212 | Yes [22] |
| Isoform 2 of CD70 Antigen (CD70) | - | - | - | - | - | - | 100 | 0.212 | Yes [23] |
| Sushi-Repeat-Containing Protein, X Chromosome (SRPX) | - | - | - | - | - | - | 100 | 0.179 | - |
| Golgi Membrane Protein 1 (GOLM1) | 100 | 0.995 | - | - | 58.319 | 0.995 | 100 | 0.166 | - |
| Stanniocalcin 1 (STC1) | - | - | - | - | - | 0.145 | 100 | 0.145 | Yes [24] |
| Transmembrane Protein 59 (TMEM59) | - | - | - | - | - | - | 100 | 0.129 | - |
| Galactosylgalactosylxylosylprotein 3-Beta-Glucuronosyltransferase 2 (B3GAT2) | - | - | - | - | - | - | 100 | 0.11 | - |
| Isoform 3 of Tyrosine-Protein Kinase Lck (LCK) | - | - | - | - | - | - | 100 | 0.08 | Yes [25] |
| Chromosome 1 Open Reading Frame 112 (C1orf112) | 1.368 | 0.061 | - | - | - | - | 100 | 0.061 | - |
| Isoform 2 of Alpha,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase B (MGAT5B) | - | - | - | - | - | - | 100 | 0.047 | - |
| Laminin Subunit Beta 2 (LAMB2) | 100 | 0.021 | - | - | 51.099 | 0.043 | 34.822 | 0.065 | - |
| Transferrin (TF) | - | - | - | - | - | - | 33.341 | 0.166 | Yes [26] |
| Carboxypeptidase (CTSA) | 3.04 | 0.096 | 1.641 | 0.905 | 2.4865 | 0.318 | 21.021 | 0.738 | Yes [3] |
| Fibrinogen-Like Protein 1 (FGL1) | - | - | - | - | - | - | 14.448 | 0.136 | - |
| Tyrosine-Protein Kinase Receptor (SDC4-ROS1_S4;R32) | - | - | - | - | 100 | 0.061 | 7.664 | 0.061 | - |
| Fructose-Bisphosphate Aldolase (ALDOC) | - | - | 0.542 | 1.593 | - | - | 6.13 | 4.736 | Yes [27] |
| Insulin-Like Growth Factor-Binding Protein 3 (IGFBP3) | - | - | - | - | 37.926 | 0.931 | 6.039 | 0.551 | Yes [28] |
| MucinB (MUC5B) | - | - | - | - | - | - | 3.878 | 0.746 | - |
| Pleckstrin Homology Domain-Containing Family B Member 2 (PLEKHB2) | - | - | - | - | - | - | 3.853 | 0.116 | - |
| Eukaryotic Initiation Factor 4A-III (EIF4A3) | 1.6715 | 13.454 | - | - | 1.5165 | 19.893 | 2.378 | 0.318 | - |
| Hepatocyte Growth Factor Receptor (MET) | 13.464 | 0.415 | - | - | 1.06 | 0.658 | 2.265 | 0.032 | Yes [29] |
| Isoform 2 Procollagen-Lysine,2-Oxoglutarate 5-Dioxygenase 2 (PLOD2) | 1.9435 | 3.885 | 0.586 | 0.848 | 10.655 | 7.149 | 2.227 | 3.642 | Yes [30] |
| Ferritin Heavy Chain (FTH1) | - | 0.359 | 1.16 | 12.594 | 8.775 | 0.166 | 2.177 | 20.544 | Yes [31] |
| Integral Membrane Proteins | Plasma Membrane Proteins | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nx EV Gene Symbols | H/L | emPAI | Hx EV Gene Symbols | H/L | emPAI | Nx EV Gene Symbols | H/L | emPAI | Hx EV Gene Symbols | H/L | emPAI | |
| CD9 | 0.531 | 26.826 | CD9 | 0.368 | 11.915 | CTSD | 11.052 | 366.47 | CTSD | 6.785 | 271.13 | |
| CD151 | 0.415 | 0.179 | CD151 | 0.269 | 0.179 | GNAS | 0.249 | 0.365 | GNAS | 0.168 | 0.283 | |
| EPHA2 | 0.152 | 0.172 | EPHA2 | 0.020 | 0.22 | LIN7A | 0.161 | 0.179 | LIN7A | 0.172 | 0.179 | |
| ATP1B1 | 0.107 | 0.311 | ATP1B1 | 0.095 | 0.311 | ARF4 | 0.233 | 0.701 | ARF4 | 0.173 | 0.701 | |
| STX5 | 0.201 | 0.585 | STX5 | 0.113 | 0.259 | CAP1 | 0.153 | 0.322 | CAP1 | 0.259 | 1.009 | |
| TSPAN9 | 0.994 | 0.425 | TSPAN9 | 0.663 | 0.194 | RAB5C | 0.321 | 1.054 | RAB5C | 0.205 | 1.738 | |
| CAV1 | 0.158 | 0.259 | CAV1 | 0.180 | 0.585 | RAB34 | 0.164 | 0.413 | RAB34 | 0.194 | 0.259 | |
| QSOX1 | 46.856 | 0.501 | QSOX1 | 15.013 | 1.581 | RAB21 | 0.265 | 0.166 | RAB21 | 0.097 | 0.166 | |
| ITGB1 | 0.126 | 1.239 | ITGB1 | 0.088 | 3.217 | RAB10 | 0.130 | 1.254 | RAB10 | 0.225 | 1.955 | |
| TSPAN15 | 0.284 | 0.389 | TSPAN15 | 0.101 | 0.179 | SDCBP | 2.380 | 128 | SDCBP | 1.567 | 17.957 | |
| NUTF2 | 0.010 | 0.334 | NUTF2 | 0.010 | 0.334 | RAB14 | 0.010 | 0.585 | RAB14 | 0.167 | 0.848 | |
| PLXNB2 | 0.010 | 0.022 | PLXNB2 | 0.025 | 0.067 | EFNB1 | 1.149 | 0.11 | EFNB1 | 0.647 | 0.233 | |
| CD81 | 0.950 | 6.943 | CD81 | 0.648 | 5.31 | RHOG | 0.160 | 0.468 | RHOG | 0.040 | 0.212 | |
| SLC16A1 | 0.135 | 0.311 | SLC16A1 | 0.061 | 0.501 | GNAI2 | 0.010 | 0.389 | GNAI2 | 0.010 | 0.551 | |
| GLUD1 | 0.092 | 0.896 | GLUD1 | 0.068 | 0.896 | RAB2A | 0.155 | 1.929 | RAB2A | 0.258 | 0.585 | |
| AXL | 3.440 | 0.179 | AXL | 2.364 | 0.245 | DSC1 | 0.010 | 0.094 | DSC1 | 0.010 | 0.094 | |
| QSOX2 | 0.230 | 0.222 | QSOX2 | 0.073 | 0.284 | RHOB | 1.273 | 0.52 | RHOB | 1.861 | 0.52 | |
| TSPAN14 | 0.850 | 4.179 | TSPAN14 | 0.661 | 2.728 | CP | 19.204 | 0.52 | CP | 11.341 | 7.111 | |
| NTRK3 | 1.113 | 0.116 | NTRK3 | 0.548 | 0.116 | RAB7A | 0.209 | 1.254 | RAB7A | 0.175 | 1.254 | |
| - | - | - | PLEKHB2 | 3.853 | 0.116 | GNA13 | 0.397 | 0.318 | GNA13 | 0.010 | 0.318 | |
| - | - | - | TPR | 0.010 | 0.016 | DSG1 | 0.010 | 0.054 | DSG1 | 0.010 | 0.054 | |
| - | - | - | MET | 2.265 | 0.032 | DMBT1 | 0.010 | 0.056 | DMBT1 | 0.010 | 0.056 | |
| - | - | - | EGFR | 0.448 | 0.033 | JUP | 0.010 | 0.17 | JUP | 0.010 | 0.17 | |
| - | - | - | PTGFRN | 0.373 | 0.136 | CD44 | 0.186 | 0.968 | CD44 | 0.084 | 0.719 | |
| GNAI3 | 0.010 | 0.438 | GNAI3 | 0.030 | 0.624 | |||||||
| RAB32 | 0.344 | 0.155 | RAB32 | 0.361 | 0.155 | |||||||
| RAB11A | 0.244 | 0.501 | - | - | - | |||||||
| TTYH3 | 100 | 0.11 | - | - | - | |||||||
| CDH1 | 2.675 | 0.194 | - | - | - | |||||||
| PLSCR1 | 0.637 | 0.52 | - | - | - | |||||||
| RHOC | 0.532 | 0.468 | - | - | - | |||||||
| SNAP23 | 0.010 | 0.179 | - | - | - | |||||||
| CDC42 | 0.254 | 0.52 | - | - | - | |||||||
| RAB8B | 0.387 | 1.054 | - | - | - | |||||||
| - | - | - | RAB11B | 0.212 | 0.778 | |||||||
| - | - | - | CLTB | 0.010 | 0.259 | |||||||
| - | - | - | CDH2 | 0.331 | 0.061 | |||||||
| - | - | - | RAB31 | 0.010 | 0.179 | |||||||
| - | - | - | GNAI1 | 0.118 | 0.833 | |||||||
| - | - | - | IGHG1 | 0.010 | 0.105 | |||||||
| - | - | - | PKP1 | 0.105 | 0.043 | |||||||
| - | - | - | CD70 | 100 | 0.212 | |||||||
| - | - | - | U3KQV3 | 0.437 | 0.166 | |||||||
| Nx EV Gene Symbol | H/L | emPAI | Hx EV Gene Symbol | H/L | emPAI |
|---|---|---|---|---|---|
| VPS35 | 0.254 | 0.047 | VPS35 | 0.140 | 0.259 |
| STX5 | 0.201 | 0.585 | STX5 | 0.113 | 0.259 |
| SDCBP | 2.380 | 128 | SDCBP | 1.567 | 17.957 |
| CAV1 | 0.158 | 0.259 | CAV1 | 0.180 | 0.585 |
| VPS29 | 0.490 | 0.778 | VPS29 | 0.261 | 1.61 |
| LMAN2 | 0.886 | 0.931 | LMAN2 | 0.403 | 0.551 |
| VPS26A | 0.370 | 0.233 | VPS26A | 0.135 | 0.369 |
| IGF2R | 0.444 | 0.418 | IGF2R | 0.344 | 1.284 |
| PDCD6IP | 0.305 | 0.874 | PDCD6IP | 0.285 | 1.125 |
| CLTC | 0.441 | 1.976 | CLTC | 0.183 | 4.867 |
| SNAP23 | 0.010 | 0.179 | - | - | - |
| SYNGR2 | 0.010 | 0.292 | - | - | - |
| - | - | - | CLTB | 0.010 | 0.259 |
| - | - | - | GOSR1 | 0.254 | 0.122 |
| - | - | - | COPA | 0.010 | 0.124 |
| - | - | - | COPE | 0.126 | 0.116 |
| - | - | - | COPB1 | 0.010 | 0.041 |
| - | - | - | COPG1 | 0.010 | 0.045 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tse, S.W.; Tan, C.F.; Park, J.E.; Gnanasekaran, J.; Gupta, N.; Low, J.K.; Yeoh, K.W.; Chng, W.J.; Tay, C.Y.; McCarthy, N.E.; et al. Microenvironmental Hypoxia Induces Dynamic Changes in Lung Cancer Synthesis and Secretion of Extracellular Vesicles. Cancers 2020, 12, 2917. https://doi.org/10.3390/cancers12102917
Tse SW, Tan CF, Park JE, Gnanasekaran J, Gupta N, Low JK, Yeoh KW, Chng WJ, Tay CY, McCarthy NE, et al. Microenvironmental Hypoxia Induces Dynamic Changes in Lung Cancer Synthesis and Secretion of Extracellular Vesicles. Cancers. 2020; 12(10):2917. https://doi.org/10.3390/cancers12102917
Chicago/Turabian StyleTse, Shun Wilford, Chee Fan Tan, Jung Eun Park, JebaMercy Gnanasekaran, Nikhil Gupta, Jee Keem Low, Kheng Wei Yeoh, Wee Joo Chng, Chor Yong Tay, Neil E. McCarthy, and et al. 2020. "Microenvironmental Hypoxia Induces Dynamic Changes in Lung Cancer Synthesis and Secretion of Extracellular Vesicles" Cancers 12, no. 10: 2917. https://doi.org/10.3390/cancers12102917
APA StyleTse, S. W., Tan, C. F., Park, J. E., Gnanasekaran, J., Gupta, N., Low, J. K., Yeoh, K. W., Chng, W. J., Tay, C. Y., McCarthy, N. E., Lim, S. K., & Sze, S. K. (2020). Microenvironmental Hypoxia Induces Dynamic Changes in Lung Cancer Synthesis and Secretion of Extracellular Vesicles. Cancers, 12(10), 2917. https://doi.org/10.3390/cancers12102917