Mass Spectrometry Imaging Reveals Neutrophil Defensins as Additional Biomarkers for Anti-PD-(L)1 Immunotherapy Response in NSCLC Patients
Abstract
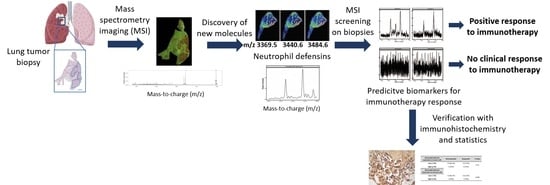
:1. Introduction
2. Results
2.1. MALDI Mass Spectrometry Imaging for Predictive Biomarker Discovery
2.2. Peptides of Interest are Identified as Neutrophil Defensin 1, 2 and 3
2.3. Neutrophil Defensins as Pretreatment Biomarkers for Immunotherapy Response
2.4. Verification of Biomarkers with Immunohistochemistry Staining (IHC)
2.5. Neutrophil Defensins Lead to In Vitro Reduced Lung Tumor Cell Growth
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Materials
4.3. Fresh Frozen Tissue Sectioning and Preparation for MSI Analysis
4.4. AMP Identification
4.5. FFPE Tissue Sectioning and Preparation for MSI Analysis
4.6. H&E Staining
4.7. Immunohistochemistry of Neutrophil Defensins
4.8. Tumor Cell Cultures, Coculturing and Cytokine (IFN-γ) Detection
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Agency for Research on Cancer. Globocan. Available online: http://Gco.Iarc.Fr/Today (accessed on 22 November 2019).
- Garon, E.B.; Hellmann, M.D.; Rizvi, N.A.; Carcereny, E.; Leighl, N.B.; Ahn, M.J.; Eder, J.P.; Balmanoukian, A.S.; Aggarwal, C.; Horn, L.; et al. Five-year overall survival for patients with advanced non-small-cell lung cancer treated with pembrolizumab: Results from the phase i KEYNOTE-001 study. J. Clin. Oncol. 2019, 37, 2518–2527. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.B.; Peng, C.; Sosman, J.A. Nivolumab in melanoma: Latest evidence and clinical potential. Ther. Adv. Med. Oncol. 2015, 7, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Teixidó, C.; Vilariño, N.; Reyes, R.; Reguart, N. PD-L1 expression testing in non-small cell lung cancer. Ther. Adv. Med. Oncol. 2018, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Riley, J.L. PD-1 signaling in primary T cells. Immunol. Rev. 2009, 229, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I.; Coukos, G.; Dranoff, G. Cancer immunotherapy comes of age. Nature 2014, 480, 480–489. [Google Scholar] [CrossRef]
- U.S. Food & Drug Administration (FDA). Available online: https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm617471.htm (accessed on 7 January 2020).
- U.S. Food & Drug Administration (FDA). Available online: https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm617370.htm (accessed on 7 January 2020).
- U.S. Food & Drug Administration (FDA). Available online: https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm525780.htm (accessed on 7 January 2020).
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef]
- Davies, M. New modalities of cancer treatment for NSCLC: Focus on immunotherapy. Cancer Manag. Res. 2014, 6, 63–75. [Google Scholar] [CrossRef] [Green Version]
- Blair, H.A. Atezolizumab: A review in previously treated advanced non-small cell lung cancer. Target. Oncol. 2018, 13, 399–407. [Google Scholar] [CrossRef]
- Tray, N.; Weber, J.S.; Adams, S. Predictive biomarkers for checkpoint immunotherapy: Current status and challenges for clinical application. Cancer Immunol. Res. 2018, 6, 1122–1128. [Google Scholar] [CrossRef] [Green Version]
- Khunger, M.; Hernandez, A.V.; Pasupuleti, V.; Rakshit, S.; Pennell, N.A.; Stevenson, J.; Mukhopadhyay, S.; Schalper, K.; Velcheti, V. Programmed cell death 1 (PD-1) ligand (PD-L1) expression in solid tumors as a predictive biomarker of benefit from PD-1/PD-L1 axis inhibitors: A systematic review and meta-analysis. JCO Precis. Oncol. 2017, 1, 1–15. [Google Scholar] [CrossRef]
- Velcheti, V.; Schalper, K.A.; Carvajal, D.E.; Rimm, D.L.; Anagnostou, V.K.; Syrigos, K.N.; Sznol, M.; Herbst, R.S.; Gettinger, S.N.; Chen, L. Programmed death ligand-1 expression in non-small cell lung cancer. Lab. Investig. 2014, 94, 107–116. [Google Scholar] [CrossRef]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2015, 20, 5064–5074. [Google Scholar] [CrossRef] [Green Version]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [Green Version]
- Daud, A.I.; Wolchok, J.D.; Robert, C.; Hwu, W.J.; Weber, J.S.; Ribas, A.; Hodi, F.S.; Joshua, A.M.; Kefford, R.; Hersey, P.; et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J. Clin. Oncol. 2016, 34, 4102–4109. [Google Scholar] [CrossRef]
- Vilain, R.E.; Menzies, A.M.; Wilmott, J.S.; Kakavand, H.; Madore, J.; Guminski, A.; Liniker, E.; Kong, B.Y.; Cooper, A.J.; Howle, J.R.; et al. Dynamic changes in PD-L1 expression and immune infiltrates early during treatment predict response to PD-1 blockade in Melanoma. Clin. Cancer Res. 2017, 23, 5024–5033. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, J.; Zwaenepoel, K.; Rolfo, C.; Van den Bossche, J.; Deben, C.; Silence, K.; Hermans, C.; Smits, E.; Van Schil, P.; Lardon, F.; et al. Unlocking the potential of CD70 as a novel immunotherapeutic target for non-small cell lung cancer. Oncotarget 2015, 6, 13462–13475. [Google Scholar] [CrossRef] [Green Version]
- Teixidó, C.; Karachaliou, N.; González-Cao, M.; Morales-Espinosa, D.; Rosell, R. Assays for predicting and monitoring responses to lung cancer immunotherapy. Cancer Biol. Med. 2015, 12, 87–95. [Google Scholar] [CrossRef]
- Berghmans, E.; Van Raemdonck, G.; Schildermans, K.; Willems, H.; Boonen, K.; Maes, E.; Mertens, I.; Pauwels, P.; Baggerman, G. MALDI mass spectrometry imaging linked with top-down proteomics as a tool to study the non-small-cell lung cancer tumor microenvironment. Methods Protoc. 2019, 2, 44. [Google Scholar] [CrossRef] [Green Version]
- Chughtai, K.; Heeren, R.M.A. Mass spectrometric imaging for biomedical tissue analysis—Chemical reviews (ACS Publications). Chem. Rev. 2011, 110, 3237–3277. [Google Scholar] [CrossRef] [Green Version]
- Ahlf Wheatcraft, D.R.; Liu, X.; Hummon, A.B. Sample preparation strategies for mass spectrometry imaging of 3D cell culture models. J. Vis. Exp. 2014, 5, e52313. [Google Scholar] [CrossRef] [Green Version]
- Minerva, L.; Clerens, S.; Baggerman, G.; Arckens, L. Direct profiling and identification of peptide expression differences in the pancreas of control and ob/ob mice by imaging mass spectrometry. Proteomics 2008, 8, 3763–3774. [Google Scholar] [CrossRef]
- Stoeckli, M.; Chaurand, P.; Hallahan, D.E.; Caprioli, R.M. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 2001, 7, 493–496. [Google Scholar] [CrossRef]
- Minerva, L.; Boonen, K.; Menschaert, G.; Landuyt, B.; Baggerman, G.; Arckens, L. Linking mass spectrometric imaging and traditional peptidomics: A validation in the obese mouse model. Anal. Chem. 2011, 83, 7682–7691. [Google Scholar] [CrossRef]
- Gaspar, D.; Freire, J.M.; Pacheco, T.R.; Barata, J.T.; Castanho, M.A.R.B. Apoptotic human neutrophil peptide-1 anti-tumor activity revealed by cellular biomechanics. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 308–316. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Wang, Y.-S.; Pan, W.-B.; Xiao, B.; Wen, Y.-J.; Chen, X.-C.; Chen, L.-J.; Deng, H.-X.; You, J.; Kan, B.; et al. Human -defensin-1 inhibits growth of human lung adenocarcinoma xenograft in nude mice. Mol. Cancer Ther. 2008, 7, 1588–1597. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.A.; Chakravarthy, A.B.; Rosenbluth, J.M.; Mi, D.; Seeley, E.H.; Granja-Ingram, N.D.M.; Olivares, M.G.; Kelley, M.C.; Mayer, I.A.; Meszoely, I.M.; et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin. Cancer Res. 2010, 16, 681–690. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Dong, H.; Li, Y.; Ma, T.; Huang, H.; Leong, H.-S.; Eckel-Passow, J.; Kocher, J.-P.A.; Liang, H.; Wang, L.; et al. Prevalent homozygous deletions of type I interferon and defensin genes in human cancers associate with immunotherapy resistance. Clin. Cancer Res. 2018, 24, 3299–3308. [Google Scholar] [CrossRef] [Green Version]
- Mukae, H.; Iiboshi, H.; Nakazato, M.; Hiratsuka, T.; Tokojima, M.; Abe, K.; Ashitani, J.; Kadota, J.; Matsukura, S.; Kohno, S. Raised plasma concentrations of α-defensins in patients with idiopathic pulmonary fibrosis. Thorax 2002, 57, 623–628. [Google Scholar] [CrossRef] [Green Version]
- Stenzinger, A.; Schwamborn, K.; Kazdal, D.; Fresnais, M.; Schirmacher, P.; Casadonte, R.; Leichsenring, J.; Kriegsmann, M.; Kriegsmann, K.; Zgorzelski, C.; et al. Combined immunohistochemistry after mass spectrometry imaging for superior spatial information. PROTEOMICS–Clin. Appl. 2018, 13, 1800035. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.A.; Markovic-Lipkovski, J.; Klatt, T.; Gamper, J.; Schwarz, G.; Beck, H.; Deeg, M.; Kalbacher, H.; Widmann, S.; Wessels, J.T.; et al. Human α-defensins HNPs-1, -2, and -3 in renal cell carcinoma. Am. J. Pathol. 2002, 160, 1311–1324. [Google Scholar] [CrossRef]
- Ferdowsi, S.; Pourfathollah, A.A.; Amiri, F.; Rafiee, M.H.; Aghaei, A. Evaluation of anticancer activity of α-defensins purified from neutrophils trapped in leukoreduction filters. Life Sci. 2019, 224, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Mothes, H.; Melle, C.; Ernst, G.; Kaufmann, R.; Von Eggeling, F.; Settmacher, U. Human neutrophil peptides 1-3—Early markers in development of colorectal adenomas and carcinomas. Dis. Markers 2008, 25, 123–129. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; Osaki, T.; Minamino, N. Large-scale identification of endogenous secretory peptides using electron transfer dissociation mass spectrometry. Mol. Cell. Proteom. 2013, 12, 700–709. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.R.; Ma, X.; Zhang, X.; Xia, Y. Electron transfer dissociation (ETD) of peptides containing intrachain disulfide bonds. J. Am. Soc. Mass Spectrom. 2012, 23, 310–320. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-L.; Jiang, H.; Lu, Q.; Dai, S.; Hancock, W.S.; Karger, B.L. Mass spectrometric determination of disulfide linkages in recombinant therapeutic proteins using on-line LC-MS with electron transfer dissociation (ETD). Anal. Chem. 2009, 81, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Compton, P.D.; Strukl, J.V.; Bai, D.L.; Shabanowitz, J.; Hunt, D.F. Optimization of electron transfer dissociation via informed selection of reagents and operating parameters. Anal. Chem. 2012, 84, 1781–1785. [Google Scholar] [CrossRef] [Green Version]
- Metz, B.; Kersten, G.F.A.; Baart, G.J.E.; De Jong, A.; Meiring, H.; Ten Hove, J.; Van Steenbergen, M.J.; Hennink, W.E.; Crommelin, D.J.A.; Jiskoot, W. Identification of formaldehyde-induced modifications in proteins: Reactions with insulin. Bioconjug. Chem. 2006, 17, 815–822. [Google Scholar] [CrossRef]
- Rahimi, F.; Shepherd, C.E.; Halliday, G.M.; Geczy, C.L.; Raftery, M.J. Antigen-epitope retrieval to facilitate proteomic analysis of formalin-fixed archival brain tissue. Anal. Chem. 2006, 78, 7216–7221. [Google Scholar] [CrossRef]
- Föll, M.C.; Fahrner, M.; Oria, V.O.; Kühs, M.; Biniossek, M.L.; Werner, M.; Bronsert, P.; Schilling, O. Reproducible proteomics sample preparation for single FFPE tissue slices using acid-labile surfactant and direct trypsinization. Clin. Proteom. 2018, 15, 11. [Google Scholar] [CrossRef] [Green Version]
- Ganz, T. The role of antimicrobial peptides in innate immunity. Integr. Comp. Biol. 2003, 43, 300–304. [Google Scholar] [CrossRef] [Green Version]
- Bhat, P.; Leggatt, G.; Waterhouse, N.; Frazer, I.H. Interferon-γ derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 2017, 8, e2836. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.; Singh, A.; Jothy, S.; Fraser, R.; Esch, F.; Solomon, S. The levels and biologic action of the human neutrophil granule peptide HP-1 in lung tumors. Peptides 1992, 13, 133–139. [Google Scholar] [CrossRef]
- Hattar, K.; Franz, K.; Ludwig, M.; Sibelius, U.; Wilhelm, J.; Lohmeyer, J.; Savai, R.; Subtil, F.S.B.; Dahlem, G.; Eul, B.; et al. Interactions between neutrophils and non-small cell lung cancer cells: Enhancement of tumor proliferation and inflammatory mediator synthesis. Cancer Immunol. Immunother. 2014, 63, 1297–1306. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Lee, D.; Lee, J.; Lee, S.; Lawler, S. Role of tumor-associated neutrophils in regulation of tumor growth in lung cancer development: A mathematical model. PLoS ONE 2019, 14, e0211041. [Google Scholar] [CrossRef]
- Lecot, P.; Sarabi, M.; Pereira Abrantes, M.; Mussard, J.; Koenderman, L.; Caux, C.; Bendriss-Vermare, N.; Michallet, M.-C. Neutrophil heterogeneity in cancer: From biology to therapies. Front. Immunol. 2019, 10, 2155. [Google Scholar] [CrossRef] [Green Version]
- BE71030031000 Biobank@UZA, Belgian Virtual Tumourbank funded by the National Cancer Plan. Available online: https://virtualtumourbank.kankerregister.org/tumourbank.aspx?url=BVT_home (accessed on 3 July 2017).
- Bemis, K.D.; Harry, A.; Eberlin, L.S.; Ferreira, C.; Van De Ven, S.M.; Mallick, P.; Stolowitz, M.; Vitek, O. Cardinal: An R package for statistical analysis of mass spectrometry-based imaging experiments. Bioinformatics 2015, 31, 2418–2420. [Google Scholar] [CrossRef] [Green Version]
- Fellers, R.T.; Greer, J.B.; Early, B.P.; Yu, X.; LeDuc, R.D.; Kelleher, N.L.; Thomas, P.M. ProSight lite: Graphical software to analyze top-down mass spectrometry data. Proteomics 2015, 15, 1235–1238. [Google Scholar] [CrossRef]
- Casadonte, R.; Caprioli, R.M. Proteomic analysis of formalin-fixed paraffin embedded tissue by MALDI imaging mass spectrometry. Nat. Protoc. 2012, 6, 1695–1709. [Google Scholar] [CrossRef] [Green Version]
- Kriegsmann, J.; Kriegsmann, M.; Casadonte, R. MALDI TOF imaging mass spectrometry in clinical pathology: A valuable tool for cancer diagnostics (review). Int. J. Oncol. 2015, 46, 893–906. [Google Scholar] [CrossRef] [Green Version]
- Kohn, E.C.; Azad, N.; Annunziata, C.; Dhamoon, A.S.; Whiteley, G. Proteomics as a tool for biomarker discovery. Dis. Markers 2007, 23, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Merlos Rodrigo, M.A.; Zitka, O.; Krizkova, S.; Moulick, A.; Adam, V.; Kizek, R. MALDI-TOF MS as evolving cancer diagnostic tool: A review. J. Pharm. Biomed. Anal. 2014, 95, 245–255. [Google Scholar] [CrossRef]







| Neutrophil Defensin 1 ACYCRIPACIAGERRYGTCIYQGRLWAFCC m/z 688.91; z = 5; Mass 3439.55 Da Retention Time 31.38 min | ||||||
|---|---|---|---|---|---|---|
| Name | Ion Type | Ion Number | Theoretical Mass | Observed Mass | Mass Difference (Da) | Mass Difference (ppm) |
| c8 | c | 8 | 894.42016 | 894.580505 | 0.160345 | 179.2725692 |
| c10 | c | 10 | 1110.51341 | 1110.714966 | 0.201556 | 181.4980334 |
| c12 | c | 12 | 1238.57198 | 1238.811157 | 0.239177 | 193.1070651 |
| c13 | c | 13 | 1367.61457 | 1367.952881 | 0.338311 | 247.3730592 |
| c17 | c | 17 | 1899.90158 | 1899.846436 | −0.055144 | −29.02466137 |
| y9 | y | 9 | 1082.478985 | 1082.668701 | 0.189716 | 175.2606772 |
| y12 | y | 12 | 1461.635565 | 1461.895508 | 0.259943 | 177.843921 |
| y16 | y | 16 | 1938.869145 | 1939.321045 | 0.4519 | 233.0740067 |
| z7 | z | 7 | 881.38018 | 881.510986 | 0.130806 | 148.4104169 |
| z8 | z | 8 | 938.40164 | 938.560364 | 0.158724 | 169.1429269 |
| z9 | z | 9 | 1066.46022 | 1066.680298 | 0.220078 | 206.3630653 |
| z10 | z | 10 | 1229.52355 | 1229.841064 | 0.317514 | 258.2414953 |
| z11 | z | 11 | 1342.60761 | 1342.919067 | 0.311457 | 231.9791707 |
| z12 | z | 12 | 1445.6168 | 1446.032227 | 0.415427 | 287.3700693 |
| z14 | z | 14 | 1603.68594 | 1603.703613 | 0.017673 | 11.02023754 |
| z15 | z | 15 | 1766.74927 | 1767.123413 | 0.374143 | 211.769155 |
| z16 | z | 16 | 1922.85038 | 1923.075928 | 0.225548 | 117.2987781 |
| Neutrophil Defensin Expression on Tumor Cells | Neutrophil Defensin Expression on Immune Cells | |||||
|---|---|---|---|---|---|---|
| Low (<2%) | High (≥2%) | p-Value | Low (<2%) | High (≥2%) | p-Value | |
| Gender | ||||||
| Female | 8 (47.1%) | 3 (37.5%) | 1 | 9 (56.3%) | 2 (22.2%) | 0.21 |
| Male | 9 (52.9%) | 5 (62.5%) | 7 (43.7%) | 7 (77.8%) | ||
| Histologic type | ||||||
| Adenocarcinoma | 13 (76.5%) | 4 (50%) | 0.22 | 12 (75%) | 5 (55.6%) | 0.33 |
| Squamous | 4 (23.5%) | 3 (37.5%) | 3 (18.7%) | 4 (44.4%) | ||
| Large cell | 1 (12.5%) | 1 (6.3%) | ||||
| Stage | ||||||
| IIb | 1 (5.9%) | 0.15 | 1 (11.1%) | 0.09 | ||
| IIIa | 2 (25%) | 2 (22.2%) | ||||
| IVa | 11 (64.7%) | 5 (62.5%) | 11 (68.8%) | 5 (55.6%) | ||
| IVb | 5 (29.4%) | 1 (12.5%) | 5 (31.2%) | 1 (11.1%) | ||
| Smoking status | ||||||
| Never-smoker | 2 (11.8%) | 1 (12.5%) | 1 | 1 (6.25%) | 2 (22.2%) | 0.53 |
| Smoker | 14 (82.3%) | 7 (87.5%) | 14 (87.5%) | 7 (77.8%) | ||
| Unknown | 1 (5.9%) | 1 (6.25%) | ||||
| Therapy response | ||||||
| Responder | 3 (17.6%) | 6 (75%) | 0.01 | 3 (18.7%) | 6 (66.7%) | 0.031 |
| Non-responder | 14 (82.4%) | 2 (25%) | 13 (81.3%) | 3 (33.3%) | ||
| Responding NSCLC Patients n = 9 (%) | Nonresponding NSCLC Patients n = 16 (%) | |
|---|---|---|
| Gender | ||
| Female | 4 (44%) | 7 (44%) |
| Male | 5 (56%) | 9 (56%) |
| Histologic type | ||
| Adenocarcinoma | 6 (67%) | 11 (69%) |
| Squamous | 2 (22%) | 5 (31%) |
| Large cell | 1 (11%) | |
| Stage | ||
| IIb | 1 (6%) | |
| IIIa | 2 (22%) | |
| IVa | 5 (56%) | 11 (69%) |
| IVb | 2 (22%) | 4 (25%) |
| Smoking status | ||
| Never-smoker | 2 (22%) | 1 (6%) |
| Smoker | 7 (78%) | 14 (88%) |
| Unknown | 1 (6%) | |
| Genetic aberrations | ||
| KRAS | 1 (11%) | 2 (13%) |
| BRAF | 1 (11%) | |
| EGFR | 2 (13%) | |
| Immunotherapy | ||
| Pembrolizumab | 7 (78%) | 7 (44%) |
| Nivolumab | 1 (11%) | 8 (50%) |
| Atezolizumab | 1 (11%) | 1 (6%) |
| Median survival | ||
| 1-year OS rate | 6 (67%) | 9 (56%) |
| 1-year PFS rate | 6 (67%) | 3 (19%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berghmans, E.; Jacobs, J.; Deben, C.; Hermans, C.; Broeckx, G.; Smits, E.; Maes, E.; Raskin, J.; Pauwels, P.; Baggerman, G. Mass Spectrometry Imaging Reveals Neutrophil Defensins as Additional Biomarkers for Anti-PD-(L)1 Immunotherapy Response in NSCLC Patients. Cancers 2020, 12, 863. https://doi.org/10.3390/cancers12040863
Berghmans E, Jacobs J, Deben C, Hermans C, Broeckx G, Smits E, Maes E, Raskin J, Pauwels P, Baggerman G. Mass Spectrometry Imaging Reveals Neutrophil Defensins as Additional Biomarkers for Anti-PD-(L)1 Immunotherapy Response in NSCLC Patients. Cancers. 2020; 12(4):863. https://doi.org/10.3390/cancers12040863
Chicago/Turabian StyleBerghmans, Eline, Julie Jacobs, Christophe Deben, Christophe Hermans, Glenn Broeckx, Evelien Smits, Evelyne Maes, Jo Raskin, Patrick Pauwels, and Geert Baggerman. 2020. "Mass Spectrometry Imaging Reveals Neutrophil Defensins as Additional Biomarkers for Anti-PD-(L)1 Immunotherapy Response in NSCLC Patients" Cancers 12, no. 4: 863. https://doi.org/10.3390/cancers12040863
APA StyleBerghmans, E., Jacobs, J., Deben, C., Hermans, C., Broeckx, G., Smits, E., Maes, E., Raskin, J., Pauwels, P., & Baggerman, G. (2020). Mass Spectrometry Imaging Reveals Neutrophil Defensins as Additional Biomarkers for Anti-PD-(L)1 Immunotherapy Response in NSCLC Patients. Cancers, 12(4), 863. https://doi.org/10.3390/cancers12040863