Coding of Glioblastoma Progression and Therapy Resistance through Long Noncoding RNAs
Abstract
:1. The Noncoding Genome
1.1. Small Noncoding RNAs
1.2. Long Noncoding RNAs
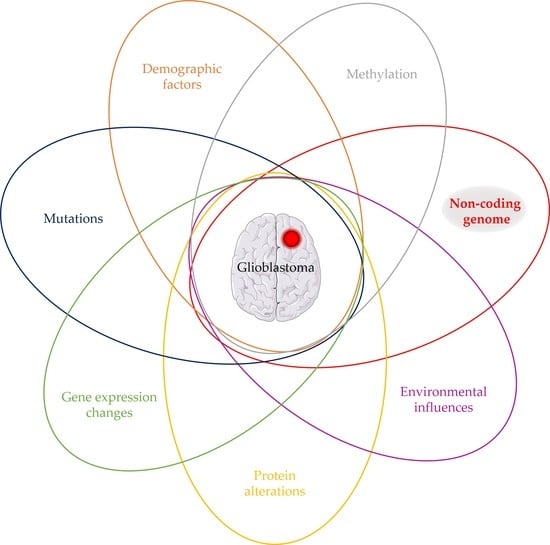
2. Glioblastoma
3. Long Noncoding RNAs and Their Relationship to Glioblastoma
3.1. Long Noncoding RNAs in Glioblastoma Subclassification and Patient Prognosis
3.2. Long Noncoding RNAs and Their Involvement in Glioblastoma Pathology
3.2.1. MALAT
3.2.2. MEG
3.2.3. HOTAIR
3.2.4. H19
3.2.5. CRNDE
4. Long Noncoding RNAs and Resistance to Glioblastoma Therapy
4.1. Temozolomide and Resistance
4.1.1. Long Noncoding RNAs Most Commonly Linked to Resistance to Glioblastoma Therapy
4.1.2. Less Common Long Noncoding RNAs Involved in Resistance to Glioblastoma Therapy
4.2. Immune-Related Long Noncoding RNAs
5. Long Noncoding RNAs and Glioblastoma Progression
5.1. Glioma Initiation
5.2. Glioma Progression
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barry, G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry 2014, 19, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Akimitsu, N. Long non-coding RNAs in cancer progression. Front. Genet. 2012, 3, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.Q.; Leung, G.K. Long non-coding RNAs in glioma: Functional roles and clinical perspectives. Neurochem. Int. 2014, 77, 78–85. [Google Scholar] [CrossRef]
- Liu, G.; Mattick, J.S.; Taft, R.J. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 2013, 12, 2061–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrescu, G.E.D.; Sabo, A.A.; Torsin, L.I.; Calin, G.A.; Dragomir, M.P. MicroRNA based theranostics for brain cancer: Basic principles. J. Exp. Clin. Cancer Res. Cr. 2019, 38, 231. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Cruickshanks, N.; Pahuski, M.; Yuan, F.; Dutta, A.; Schiff, D.; Purow, B.; Abounader, R. Noncoding RNAs in Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017. [Google Scholar] [CrossRef]
- Anglicheau, D.; Muthukumar, T.; Suthanthiran, M. MicroRNAs: Small RNAs with big effects. Transplantation 2010, 90, 105–112. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25. [Google Scholar] [CrossRef]
- Tan, W.; Liu, B.; Qu, S.; Liang, G.; Luo, W.; Gong, C. MicroRNAs and cancer: Key paradigms in molecular therapy. Oncol. Lett. 2018, 15, 2735–2742. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal. Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qureshi, I.A.; Mehler, M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012, 13, 528–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Dong, B.; Cao, J.; Mao, Y.; Guan, W.; Peng, Y.; Wang, S. Long non-coding RNA in glioma: Signaling pathways. Oncotarget 2017, 8, 27582–27592. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Xu, Z.; Li, Z.; Sun, L.; Gong, Z. An Insight into the Increasing Role of LncRNAs in the Pathogenesis of Gliomas. Front. Mol. Neurosci. 2017, 10, 53. [Google Scholar] [CrossRef] [Green Version]
- Liz, J.; Esteller, M. lncRNAs and microRNAs with a role in cancer development. Biochim. Biophys. Acta 2016, 1859, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhu, Y.; Wang, H.; Ji, X. Targeting Long Noncoding RNA in Glioma: A Pathway Perspective. Mol. Ther. Nucleic Acids 2018, 13, 431–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, T.; Li, L.; Zhou, Y.; Gao, L. Exploring Long Noncoding RNAs in Glioblastoma: Regulatory Mechanisms and Clinical Potentials. Int. J. Genom. 2018, 2018, 2895958. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Zhang, K.; Shi, Z.; Zhang, J.; Zhu, J.; Zhu, S.; Zhang, A.; Jia, Z.; Wang, G.; Yu, S.; et al. LncRNA pro fi le of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int. J. Oncol. 2012, 40, 2004–2012. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Sun, S.; Pu, J.K.; Tsang, A.C.; Lee, D.; Man, V.O.; Lui, W.M.; Wong, S.T.; Leung, G.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012, 48, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Ren, Z.; Sun, P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell. Biochem. 2012, 113, 1868–1874. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Wang, Z.; Zhou, D. Long non-coding RNAs as potential biomarkers and therapeutic targets for gliomas. Med. Hypotheses 2013, 81, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.K.; Chang, C.N.; Tsai, C.S.; Huang, Y.C.; Lu, Y.J.; Chen, W.J.; Lin, Y.H.; Chung, I.H.; Lin, K.H. The long non-coding RNA LOC441204 enhances cell growth in human glioma. Sci. Rep. 2017, 7, 5603. [Google Scholar] [CrossRef] [Green Version]
- Chamberlain, M.C. Bevacizumab for the treatment of recurrent glioblastoma. Clin. Med. Insights. Oncol. 2011, 5, 117–129. [Google Scholar] [CrossRef] [Green Version]
- Aum, D.J.; Kim, D.H.; Beaumont, T.L.; Leuthardt, E.C.; Dunn, G.P.; Kim, A.H. Molecular and cellular heterogeneity: The hallmark of glioblastoma. Neurosurg. Focus 2014, 37, E11. [Google Scholar] [CrossRef] [Green Version]
- Soeda, A.; Hara, A.; Kunisada, T.; Yoshimura, S.; Iwama, T.; Park, D.M. The evidence of glioblastoma heterogeneity. Sci. Rep. 2015, 5, 7979. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.; Yu, Y.; Grimmer, M.R.; Wahl, M.; Chang, S.M.; Costello, J.F. Temozolomide-associated hypermutation in gliomas. Neuro-Oncology 2018, 20, 1300–1309. [Google Scholar] [CrossRef] [Green Version]
- Muscat, A.M.; Wong, N.C.; Drummond, K.J.; Algar, E.M.; Khasraw, M.; Verhaak, R.; Field, K.; Rosenthal, M.A.; Ashley, D.M. The evolutionary pattern of mutations in glioblastoma reveals therapy-mediated selection. Oncotarget 2018, 9, 7844–7858. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cazzato, E.; Ladewig, E.; Frattini, V.; Rosenbloom, D.I.; Zairis, S.; Abate, F.; Liu, Z.; Elliott, O.; Shin, Y.J.; et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016, 48, 768–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sims, J.S.; Ung, T.H.; Neira, J.A.; Canoll, P.; Bruce, J.N. Biomarkers for glioma immunotherapy: The next generation. J. Neurooncol. 2015, 123, 359–372. [Google Scholar] [CrossRef]
- Lieberman, N.A.P.; Vitanza, N.A.; Crane, C.A. Immunotherapy for brain tumors: Understanding early successes and limitations. Expert Rev. Neurother. 2018, 18, 251–259. [Google Scholar] [CrossRef]
- Artene, S.A.; Tuta, C.; Dragoi, A.; Alexandru, O.; Stefana Oana, P.; Tache, D.E.; Danciulescu, M.M.; Boldeanu, M.V.; Silosi, C.A.; Dricu, A. Current and emerging EGFR therapies for glioblastoma. J. Immunoass. Immunochem 2018, 39, 1–11. [Google Scholar] [CrossRef]
- Paolillo, M.; Boselli, C.; Schinelli, S. Glioblastoma under Siege: An Overview of Current Therapeutic Strategies. Brain Sci. 2018, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Bolouri, H.; Zhao, L.P.; Holland, E.C. Big data visualization identifies the multidimensional molecular landscape of human gliomas. Proc. Natl. Acad. Sci. USA 2016, 113, 5394–5399. [Google Scholar] [CrossRef] [Green Version]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Wang, X.P.; Shan, C.; Deng, X.L.; Li, L.Y.; Ma, W. Long non-coding RNA PAR5 inhibits the proliferation and progression of glioma through interaction with EZH2. Oncol. Rep. 2017, 38, 3177–3186. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Liu, C.; Wu, M. New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer 2018, 17, 61. [Google Scholar] [CrossRef] [PubMed]
- Kiang, K.M.; Zhang, X.Q.; Leung, G.K. Long Non-Coding RNAs: The Key Players in Glioma Pathogenesis. Cancers (Basel) 2015, 7, 1406–1424. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.Q.; Sun, S.; Lam, K.F.; Kiang, K.M.; Pu, J.K.; Ho, A.S.; Lui, W.M.; Fung, C.F.; Wong, T.S.; Leung, G.K. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol. Dis. 2013, 58, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qian, J.; Wang, Y.Y.; Zhang, J.X.; You, Y.P. Long noncoding RNA profiles reveal three molecular subtypes in glioma. CNS Neurosci. Ther. 2014, 20, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Kraus, T.F.; Greiner, A.; Guibourt, V.; Lisec, K.; Kretzschmar, H.A. Identification of Stably Expressed lncRNAs as Valid Endogenous Controls for Profiling of Human Glioma. J. Cancer 2015, 6, 111–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wang, P.; Wang, Y.; Ma, X.; Zhi, H.; Zhou, D.; Li, X.; Fang, Y.; Shen, W.; Xu, Y.; et al. Lnc2Cancer v2.0: Updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2019, 47, D1028–D1033. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Zhang, J.; Wang, P.; Zhi, H.; Wang, J.; Liu, Y.; Gao, Y.; Guo, M.; Yue, M.; Wang, L.; et al. Lnc2Cancer: A manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016, 44, D980–D985. [Google Scholar] [CrossRef]
- Gravendeel, L.A.; Kouwenhoven, M.C.; Gevaert, O.; de Rooi, J.J.; Stubbs, A.P.; Duijm, J.E.; Daemen, A.; Bleeker, F.E.; Bralten, L.B.; Kloosterhof, N.K.; et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009, 69, 9065–9072. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Hui, A.M.; Su, Q.; Vortmeyer, A.; Kotliarov, Y.; Pastorino, S.; Passaniti, A.; Menon, J.; Walling, J.; Bailey, R.; et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006, 9, 287–300. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.T.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Wu, Z.; Wu, T.; Huang, Y.; Cheng, Z.; Li, X.; Sun, T.; Xie, X.; Zhou, Y.; Du, Z. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, J.P.; Guo, S.F.; Jiang, S.L.; Xu, Y.L.; Li, J.W.; Li, L.; Xiang, J.Y. Silencing of Long Non-Coding RNA MALAT1 Promotes Apoptosis of Glioma Cells. J. Korean Med. Sci. 2016, 31, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Xu, C.Y.; Ding, B.Q.; Gao, M.; Wei, X.T.; Ji, N. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J. Neurooncol. 2017, 134, 19–28. [Google Scholar] [CrossRef]
- Xiong, Z.Y.; Wang, L.Y.; Wang, Q.P.; Yuan, Y. LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J. Cell. Mol. Med. 2018, 22, 3929–3940. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.K.; Wang, Y.L.; Li, H.; Sun, Y.; Gong, Y.H. lncRNAs combine and crosstalk with NSPc1 in ATRA-induced differentiation of U87 glioma cells. Oncol. Lett. 2019, 17, 5821–5829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Liu, J.; Quan, J.; Liu, W.L.; Tan, H.; Li, W.P. lncRNAs as potential molecular biomarkers for the clinicopathology and prognosis of glioma: A systematic review and meta-analysis. Gene 2018, 668, 77–86. [Google Scholar] [CrossRef]
- Ma, K.X.; Wang, H.J.; Li, X.R.; Li, T.; Su, G.; Yang, P.; Wu, J.W. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumor Biol. 2015, 36, 3355–3359. [Google Scholar] [CrossRef]
- Wylie, A.A.; Murphy, S.K.; Orton, T.C.; Jirtle, R.L. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000, 10, 1711–1718. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, N.; Wagatsuma, H.; Wakana, S.; Shiroishi, T.; Nomura, M.; Aisaka, K.; Kohda, T.; Surani, M.A.; Kaneko-Ishino, T.; Ishino, F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 2000, 5, 211–220. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Y.; Mehta, K.R.; Danila, D.C.; Scolavino, S.; Johnson, S.R.; Klibanski, A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 2003, 88, 5119–5126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Wang, X.; Feng, X.; Li, X.; Pan, L.; Liu, J.; Wang, F.; Yuan, Z.; Yang, L.; Yu, J.; et al. Long non-coding RNA MEG3 regulates proliferation, apoptosis, and autophagy and is associated with prognosis in glioma. J. Neurooncl. 2018, 140, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Yu, H.; Liu, X.; Liu, Y.; Zheng, J.; Wang, P.; Gong, W.; Chen, J.; Zhao, L.; Xue, Y. PIWIL1/piRNA-DQ593109 Regulates the Permeability of the Blood-Tumor Barrier via the MEG3/miR-330-5p/RUNX3 Axis. Mol. Ther. Nucleic Acids 2018, 10, 412–425. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.D.; Guo, W.S. Long non-coding RNA MEG3 suppresses the growth of glioma cells by regulating the miR-96-5p/MTSS1 signaling pathway. Mol. Med. Rep. 2019, 20, 4215–4225. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.H.; Chi, G.N.; Zhao, C.H.; Li, D.Y. Long noncoding RNA MEG3 inhibits proliferation and migration but induces autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in glioma cells. J. Cell. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fu, C.W.; Fan, D.Q. Participation of tumor suppressors long non-coding RNA MEG3, microRNA-377 and PTEN in glioma cell invasion and migration. Pathol. Res. Pract. 2019, 215. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, X.; Li, Y.X. Long non-coding RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and inactivating PI3K/AKT pathway. Oncol. Rep. 2017, 38, 2408–2416. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, K.; Luo, Z.; Liu, L.; Wu, L.; Liu, J. Circulating long non-coding HOX transcript antisense intergenic ribonucleic acid in plasma as a potential biomarker for diagnosis of breast cancer. Thorac. Cancer 2016, 7, 627–632. [Google Scholar] [CrossRef]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Song, X.; Wang, X.; Xie, Y.; Wang, Z.; Xu, Y.; You, X.; Liang, Z.; Cao, H. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res. Treat. 2015, 152, 199–208. [Google Scholar] [CrossRef]
- Wang, W.; He, X.; Zheng, Z.; Ma, X.; Hu, X.; Wu, D.; Wang, M. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol. Cancer 2017, 16, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastori, C.; Kapranov, P.; Penas, C.; Peschansky, V.; Volmar, C.H.; Sarkaria, J.N.; Bregy, A.; Komotar, R.; St Laurent, G.; Ayad, N.G.; et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. USA 2015, 112, 8326–8331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.K.; Pastori, C.; Penas, C.; Komotar, R.J.; Ivan, M.E.; Wahlestedt, C.; Ayad, N.G. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer 2018, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.-X.; Han, L.; Bao, Z.-S.; Wang, Y.-Y.; Chen, L.-Y.; Yan, W.; Yu, S.-Z.; Pu, P.-Y.; Liu, N.; You, Y.-P.; et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma (vol 15, pg 1595, 2013). Neuro-Oncology 2017, 19, 1703. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.H.; Yuan, H.Y.; Ren, X.Y.; Huang, K.; Guo, Z.Y. Association between expression of HOTAIR and invasiveness of gliomas, and its predictive value. Adv. Clin. Exp. Med. 2019, 28, 1179–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xavier-Magalhaes, A.; Goncalves, C.S.; Fogli, A.; Lourenco, T.; Pojo, M.; Pereira, B.; Rocha, M.; Lopes, M.C.; Crespo, I.; Rebelo, O.; et al. The long non-coding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma. Oncotarget 2018, 9, 15740–15756. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Sun, X.; Zhou, X.; Han, L.; Chen, L.; Shi, Z.; Zhang, A.; Ye, M.; Wang, Q.; Liu, C.; et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget 2015, 6, 537–546. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Tycko, B. Monoallelic expression of the human H19 gene. Nat. Genet. 1992, 1, 40–44. [Google Scholar] [CrossRef]
- Ariel, I.; de Groot, N.; Hochberg, A. Imprinted H19 gene expression in embryogenesis and human cancer: The oncofetal connection. Am. J. Med Genet. 2000, 91, 46–50. [Google Scholar] [CrossRef]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef]
- Li, W.; Jiang, P.; Sun, X.; Xu, S.; Ma, X.; Zhan, R. Suppressing H19 Modulates Tumorigenicity and Stemness in U251 and U87MG Glioma Cells. Cell. Mol. Neurobiol. 2016, 36, 1219–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Yan, Y.; Hu, M.; Chen, X.; Wang, Y.; Dai, Y.; Wu, D.; Wang, Y.; Zhuang, Z.; Xia, H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J. Neurosurg. 2016, 2016, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, Y.R.; Zeng, F.; Cao, H.Y.; Zhou, H.D.; Wang, Y.J. LncRNA H19 is overexpressed in glioma tissue, is negatively associated with patient survival, and promotes tumor growth through its derivative miR-675. Eur. Rev. Med Pharmacol. Sci. 2016, 20, 4891–4897. [Google Scholar]
- Jia, P.; Cai, H.; Liu, X.; Chen, J.; Ma, J.; Wang, P.; Liu, Y.; Zheng, J.; Xue, Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016, 381, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Peng, R.; Liu, Q.; Liu, D.; Du, P.; Yuan, J.; Peng, G.; Liao, Y. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch. Biochem. Biophys. 2016, 610, 1–7. [Google Scholar] [CrossRef]
- Liu, Z.Z.; Tian, Y.F.; Wu, H.; Ouyang, S.Y.; Kuang, W.L. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1alpha/VEGF axis. Neoplasma 2019. [Google Scholar] [CrossRef]
- Li, C.; Lei, B.; Huang, S.; Zheng, M.; Liu, Z.; Li, Z.; Deng, Y. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am. J. Transl. Res. 2015, 7, 1747–1764. [Google Scholar]
- Shi, Y.; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS ONE 2014, 9, e86295. [Google Scholar] [CrossRef] [Green Version]
- Guan, N.; Wang, R.; Guo, W.S.; Lai, Y.J.; Zhang, Y.D.; Cheng, Y.Y. Long non-coding RNA H19 regulates the development of gliomas through the Wnt/beta-catenin signaling pathway. Eur. Rev. Med Pharmacol. Sci. 2019, 23, 4243–4253. [Google Scholar]
- Wu, W.; Hu, Q.; Nie, E.; Yu, T.; Wu, Y.; Zhi, T.; Jiang, K.; Shen, F.; Wang, Y.; Zhang, J.; et al. Hypoxia induces H19 expression through direct and indirect Hif-1alpha activity, promoting oncogenic effects in glioblastoma. Sci. Rep. 2017, 7, 45029. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Yin, J.; Zeng, A.; Jin, X.; Zhang, Z.; Yan, W.; You, Y. H19 Functions as a Competing Endogenous RNA to Regulate EMT by Sponging miR-130a-3p in Glioma. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 50, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Graham, L.D.; Pedersen, S.K.; Brown, G.S.; Ho, T.; Kassir, Z.; Moynihan, A.T.; Vizgoft, E.K.; Dunne, R.; Pimlott, L.; Young, G.P.; et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer 2011, 2, 829–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szafron, L.M.; Balcerak, A.; Grzybowska, E.A.; Pienkowska-Grela, B.; Podgorska, A.; Zub, R.; Olbryt, M.; Pamula-Pilat, J.; Lisowska, K.M.; Grzybowska, E.; et al. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget 2015, 6, 43897–43910. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.L.; Wang, Y.T.; Li, J.F.; Zhang, Y.Z.; Yin, H.L.; Han, B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015, 367, 122–128. [Google Scholar] [CrossRef]
- Li, D.X.; Fei, X.R.; Dong, Y.F.; Cheng, C.D.; Yang, Y.; Deng, X.F.; Huang, H.L.; Niu, W.X.; Zhou, C.X.; Xia, C.Y.; et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget 2017, 8, 88163–88178. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Li, X.D.; Wang, P.; Liu, X.B.; Xue, Y.X.; Hu, Y.; Li, Z.; Li, Z.Q.; Wang, Z.H.; Liu, Y.H. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget 2015, 6, 25339–25355. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Liu, X.; Wang, P.; Xue, Y.; Ma, J.; Qu, C.; Liu, Y. CRNDE Promotes Malignant Progression of Glioma by Attenuating miR-384/PIWIL4/STAT3 Axis. Mol. Ther. J. Am. Soc. Gene Ther. 2016, 24, 1199–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, S.Y.; Lu, Y.Y.; Yang, J.K.; Deng, W.Y.; Zhou, Q.; Jiao, B.H. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3992–3996. [Google Scholar]
- Kiang, K.M.; Zhang, X.Q.; Zhang, G.P.; Li, N.; Cheng, S.Y.; Poon, M.W.; Pu, J.K.; Lui, W.M.; Leung, G.K. CRNDE Expression Positively Correlates with EGFR Activation and Modulates Glioma Cell Growth. Target. Oncol. 2017, 12, 353–363. [Google Scholar] [CrossRef]
- Li, H.; Li, Q.; Guo, T.; He, W.; Dong, C.; Wang, Y. LncRNA CRNDE triggers inflammation through the TLR3-NF-kappaB-Cytokine signaling pathway. Tumour Biol. J. Int. Soc. Oncodevelop. Biol. Med. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Strobel, H.; Baisch, T.; Fitzel, R.; Schilberg, K.; Siegelin, M.D.; Karpel-Massler, G.; Debatin, K.M.; Westhoff, M.A. Temozolomide and Other Alkylating Agents in Glioblastoma Therapy. Biomedicines 2019, 7, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meir, E.G.; Hadjipanayis, C.G.; Norden, A.D.; Shu, H.K.; Wen, P.Y.; Olson, J.J. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J. Clin. 2010, 60, 166–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xu, X.K.; Li, J.L.; Kong, K.K.; Li, H.; Chen, C.; He, J.; Wang, F.; Li, P.; Ge, X.S.; et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget 2017, 8, 22783–22799. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Yuan, X.; Yan, D.; Li, D.; Guan, F.; Dong, Y.; Wang, H.; Liu, X.; Yang, B. Long Non-Coding RNA MALAT1 Decreases the Sensitivity of Resistant Glioblastoma Cell Lines to Temozolomide. Cell Physiol. Biochem. 2017, 42, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Cai, T.; Liu, Y.; Xiao, J. Long noncoding RNA MALAT1 knockdown reverses chemoresistance to temozolomide via promoting microRNA-101 in glioblastoma. Cancer Med. 2018, 7, 1404–1415. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.; Tian, Y.; Chen, Y.; Zhang, G. The silencing of LncRNA-H19 decreases chemoresistance of human glioma cells to temozolomide by suppressing epithelial-mesenchymal transition via the Wnt/beta-Catenin pathway. Oncotargets Ther. 2018, 11, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Wang, P.; Sun, X.; Yuan, Z.; Zhan, R.; Ma, X.; Li, W. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. Oncotargets Ther. 2016, 9, 3501–3509. [Google Scholar] [CrossRef] [Green Version]
- Shang, C.; Tang, W.; Pan, C.; Hu, X.; Hong, Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother. Pharm. 2018, 81, 671–678. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, N.; Liu, B.; Huang, Y.; Zeng, H.; Yang, Z.; He, Z.; Guo, H. Long noncoding RNA RP11-838N2.4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget 2016, 7, 43835–43851. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Gao, Y.; Guo, R.; Li, H.; Yang, B. Microarray expression profiles and bioinformatics analysis of mRNAs, lncRNAs, and circRNAs in the secondary temozolomide-resistant glioblastoma. Invest. N. Drugs 2019. [Google Scholar] [CrossRef]
- Wu, P.; Cai, J.; Chen, Q.; Han, B.; Meng, X.; Li, Y.; Li, Z.; Wang, R.; Lin, L.; Duan, C.; et al. Lnc-TALC promotes O(6)-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat. Commun. 2019, 10, 2045. [Google Scholar] [CrossRef]
- Yan, Y.; Xu, Z.; Chen, X.; Wang, X.; Zeng, S.; Zhao, Z.; Qian, L.; Li, Z.; Wei, J.; Huo, L.; et al. Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front Cell Dev. Biol. 2019, 7, 217. [Google Scholar] [CrossRef] [Green Version]
- Mazor, G.; Levin, L.; Picard, D.; Ahmadov, U.; Caren, H.; Borkhardt, A.; Reifenberger, G.; Leprivier, G.; Remke, M.; Rotblat, B. The lncRNA TP73-AS1 is linked to aggressiveness in glioblastoma and promotes temozolomide resistance in glioblastoma cancer stem cells. Cell Death Dis. 2019, 10, 246. [Google Scholar] [CrossRef]
- Xu, N.; Liu, B.; Lian, C.; Doycheva, D.M.; Fu, Z.; Liu, Y.; Zhou, J.; He, Z.; Yang, Z.; Huang, Q.; et al. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis 2018, 9, 1139. [Google Scholar] [CrossRef]
- Zhang, Z.; Yin, J.; Lu, C.; Wei, Y.; Zeng, A.; You, Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 166. [Google Scholar] [CrossRef]
- Chin, C.; Lunking, E.S.; de la Fuente, M.; Ayad, N.G. Immunotherapy and Epigenetic Pathway Modulation in Glioblastoma Multiforme. Front. Oncol. 2018, 8, 521. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Chen, J.; Yang, L.; Ouyang, Q.; Li, J.; Lao, L.; Zhao, J.; Liu, J.; Lu, Y.; Xing, Y.; et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat. Immunol. 2018, 19, 1112–1125. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, J.; Yu, J.; Liu, X.; Yu, C.; Hu, J.; Shi, H.; Ma, X. Long Non-coding RNAs: Emerging Roles in the Immunosuppressive Tumor Microenvironment. Front. Oncol. 2020, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Meng, Y. Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer 2019, 19, 813. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, Z.; Zhao, H.; Bao, S.; Cheng, L.; Sun, J. An Immune-Related Six-lncRNA Signature to Improve Prognosis Prediction of Glioblastoma Multiforme. Mol. Neurobiol. 2018, 55, 3684–3697. [Google Scholar] [CrossRef]
- Xia, P.; Li, Q.; Wu, G.; Huang, Y. An Immune-Related lncRNA Signature to Predict Survival In Glioma Patients. Cell. Mol. Neurobiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, J.J.; Lin, X.B.; Bao, Y.; Chen, Z.H.; Zhang, C.R.; Cai, Z.; Zhou, J.Y.; Ding, M.H.; Wu, X.J.; et al. Differential lncRNA expression profiles in recurrent gliomas compared with primary gliomas identified by microarray analysis. Int. J. Clin. Exp. Med. 2015, 8, 5033–5043. [Google Scholar] [PubMed]
- Wang, W.; Yang, F.; Zhang, L.; Chen, J.; Zhao, Z.; Wang, H.; Wu, F.; Liang, T.; Yan, X.; Li, J.; et al. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget 2016, 7, 77225–77236. [Google Scholar] [CrossRef] [Green Version]
- Zhi, F.; Wang, Q.; Xue, L.; Shao, N.Y.; Wang, R.; Deng, D.N.; Wang, S.N.; Xia, X.W.; Yang, Y.L. The Use of Three Long Non-Coding RNAs as Potential Prognostic Indicators of Astrocytoma. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Zhen, L.; Yun-Hui, L.; Hong-Yu, D.; Jun, M.; Yi-Long, Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 2016, 37, 673–683. [Google Scholar] [CrossRef]
- Lv, Q.L.; Hu, L.; Chen, S.H.; Sun, B.; Fu, M.L.; Qin, C.Z.; Qu, Q.; Wang, G.H.; He, C.J.; Zhou, H.H. A Long Noncoding RNA ZEB1-AS1 Promotes Tumorigenesis and Predicts Poor Prognosis in Glioma. In.t J. Mol. Sci. 2016, 17, 1431. [Google Scholar] [CrossRef]
- Yan, H.; Tian, R.; Zhang, M.; Wu, J.; Ding, M.; He, J. High expression of long noncoding RNA HULC is a poor predictor of prognosis and regulates cell proliferation in glioma. OncoTargets Ther. 2017, 10, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Lv, Q.L.; Chen, S.H.; Sun, B.; Qu, Q.; Cheng, L.; Guo, Y.; Zhou, H.H.; Fan, L. Up-Regulation of Long Non-Coding RNA AB073614 Predicts a Poor Prognosis in Patients with Glioma. Int J Env. Res Public Health 2016, 13, 433. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, J.; Liu, Y.; Zhang, W.; Zhou, J.; Duan, R.; Pu, P.; Kang, C.; Han, L. A novel cell cycle-associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016, 373, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, D.L.; Pang, Q. Long noncoding RNA SPRY4-IT1 is a prognostic factor for poor overall survival and has an oncogenic role in glioma. Eur. Rev. Med. Pharm. Sci. 2016, 20, 3035–3039. [Google Scholar]
- Lv, Q.L.; Chen, S.H.; Zhang, X.; Sun, B.; Hu, L.; Qu, Q.; Huang, Y.T.; Wang, G.H.; Liu, Y.L.; Zhang, Y.Y.; et al. Upregulation of long noncoding RNA zinc finger antisense 1 enhances epithelial-mesenchymal transition in vitro and predicts poor prognosis in glioma. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Zhang, C.; Cai, J.; Yang, F.; Liang, T.; Yan, X.; Wang, H.; Wang, W.; Chen, J.; Jiang, T. Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma. Oncotarget 2017, 8, 53110–53123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Li, Y.; Zhu, G.; Tian, B.; Zeng, W.; Yang, Y.; Li, Z. Long noncoding RNA CASC2 predicts the prognosis of glioma patients and functions as a suppressor for gliomas by suppressing Wnt/beta-catenin signaling pathway. Neuropsychiatr. Dis. Treat. 2017, 13, 1805–1813. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.Y.; Yao, J.; Geng, P.L.; Fu, X.P.; Xue, J.H.; Zhang, Z.W. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Patho. 2014, 7, 3065–3072. [Google Scholar]
- Shang, C.; Guo, Y.; Hong, Y.; Xue, Y.X. Long Non-coding RNA TUSC7, a Target of miR-23b, Plays Tumor-Suppressing Roles in Human Gliomas. Front Cell Neurosci. 2016, 10, 235. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Xue, Y.; Wang, P.; Wang, Z.; Li, Z.; Hu, Y.; Li, Z.; Shang, X.; Liu, Y. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget 2015, 6, 19759–19779. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, M.; An, G.; Ma, Q.F. LncRNA TUG1 acts as a tumor suppressor in human glioma by promoting cell apoptosis. Exp. Biol. Med. 2016, 241, 644–649. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; An, G.; Zhang, M.; Ma, Q. Long non-coding RNA TUG1 acts as a miR-26a sponge in human glioma cells. Biochem. Biophys. Res. Commun. 2016, 477, 743–748. [Google Scholar] [CrossRef]
- Li, Z.; Tan, H.; Zhao, W.; Xu, Y.; Zhang, Z.; Wang, M.; Zhou, X. Integrative analysis of DNA methylation and gene expression profiles identifies MIR4435-2HG as an oncogenic lncRNA for glioma progression. Gene 2019, 715, 144012. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wang, S.; Fritah, S.; Wang, X.; Zhou, W.; Yang, N.; Ni, S.; Huang, B.; Chen, A.; Li, G.; et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/beta-catenin signalling. Brain 2019. [Google Scholar] [CrossRef] [Green Version]
- Ramos, A.D.; Attenello, F.J.; Lim, D.A. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci. Lett. 2016, 625, 70–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| lncRNA Up- (↑)/Down- (↓) Regulation in TMZ-Resistant Model | Function in Context of Resistance | Model | Reference |
|---|---|---|---|
| ↓ NONHSAT163779 ↓ NONHSAT170564 ↓ lnc-NRK-1:2 ↓ lnc-MAP3K9-8:1 ↓ lnc-LOXL3-4:1 ↓ NONHSAT178713 ↓ lnc-COX11-5:1 ↓ ENST00000420774 ↑ NONHSAT154798 ↓ lnc-MYC-16:1 ↓ NONHSAT171282 ↓ T277882 ↑ NONHSAT218984 ↑ NONHSAT211205 ↑ NONHSAT186818 ↓ NR_037403 ↑ T038545 ↑ T350149 ↑ NONHSAT178873 ↑ NONHSAT170013 | NONHSAT163779 regulates ALG13, an enzyme involved in glycosylation | Secondary TMZ-resistant glioblastoma | Zhao et al. |
| ↑ H19 | Promotes chemoresistance through EMT and Wnt/ß-catenin pathway | TMZ-resistant U251 and M059J | Jia et al. |
| ↑ H19 | Linked to overexpression of drug-resistance genes | Glioblastoma tissue from patients TMZ-resistant U87 and U251 | Jiang et al. |
| ↑ MALAT1 | Regulation thymidylate synthase expression via miR-203 | Glioblastoma tissue and serum from patients TMZ-resistant U87 and U251 | Chen et al. |
| ↑ MALAT1 | Linked to the expression of multidrug-resistance proteins and EMT biomarkers | TMZ-resistant U87 and U251 | Li et al. |
| ↑ MALAT1 | Regulates miR-101 | TMZ-resistant U251 | Cai et al. |
| ↓ TUSC7 | Regulated MDR1 via binding to miR-10a | Glioblastoma tissue from patients TMZ-resistant U87 glioblastoma | Shang et al. |
| ↓ RP11-838N2.4 | Regulates EphA8 via miR-10a Regulates TGF-β signaling pathway independent of miR-10a | Glioblastoma tissue from patients TMZ-resistant U251 and U87 | Liu et al. |
| ↑ TALC | Promotes MGMT through regulation of c-Met via mIR-20b-3p | TMZ-resistant LN229, U251, 551W, and HG7 | Wu et al. |
| ↑ ADAMTS9-AS2 | Binding FUS protein and enhances its stability | Tissue from glioblastoma patients TMZ-resistant T98G and U118 | Yan et al. |
| ↑ TP73-AS1 | Regulates ALDH1A1 | Glioblastoma tissue G26, G7—glioblastoma stem cells | Mazor et al. |
| ↓ AC003092.1 | Regulates TFPI-2 via miR-195 | Glioblastoma tissue patients TMZ-resistant U87 cell line | Xu et al. |
| ↑ SBF2-AS1 | Regulates XRCC4 via miR-151a-3p | U87, LN229, A172, T98, U251, N3, Pri GBM, N3S, Rec GBM, N3T3rd | Zhang et al. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zottel, A.; Šamec, N.; Videtič Paska, A.; Jovčevska, I. Coding of Glioblastoma Progression and Therapy Resistance through Long Noncoding RNAs. Cancers 2020, 12, 1842. https://doi.org/10.3390/cancers12071842
Zottel A, Šamec N, Videtič Paska A, Jovčevska I. Coding of Glioblastoma Progression and Therapy Resistance through Long Noncoding RNAs. Cancers. 2020; 12(7):1842. https://doi.org/10.3390/cancers12071842
Chicago/Turabian StyleZottel, Alja, Neja Šamec, Alja Videtič Paska, and Ivana Jovčevska. 2020. "Coding of Glioblastoma Progression and Therapy Resistance through Long Noncoding RNAs" Cancers 12, no. 7: 1842. https://doi.org/10.3390/cancers12071842
APA StyleZottel, A., Šamec, N., Videtič Paska, A., & Jovčevska, I. (2020). Coding of Glioblastoma Progression and Therapy Resistance through Long Noncoding RNAs. Cancers, 12(7), 1842. https://doi.org/10.3390/cancers12071842