Renal Cell Tumors: Uncovering the Biomarker Potential of ncRNAs
Abstract
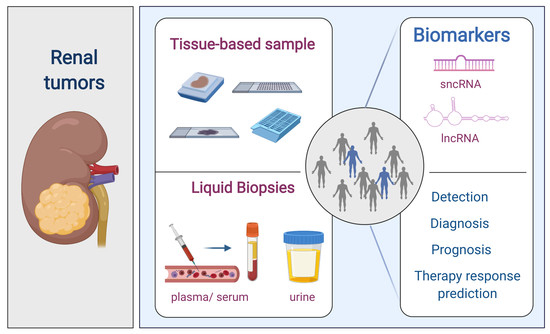
:1. Renal Cell Tumors
2. Epigenetics
3. Evidence Acquisition
4. Non-Coding RNAs (ncRNAs)
4.1. Small Non-Coding RNAs (sncRNAs)
4.1.1. MiRNAs and piRNAs in RCTs
4.1.2. SncRNAs as Diagnostic Biomarkers
Tissue-Based Samples
Liquid Biopsies
4.1.3. SncRNAs as Prognostic Biomarkers
Tissue-Based Samples
Liquid Biopsies
4.1.4. SncRNAs as Predictive Biomarkers of Response to Therapy
Tissue-Based Samples
Liquid Biopsies
In Vitro Studies
4.2. LncRNAs
4.2.1. LncRNAs in RCT
4.2.2. LncRNAs as Diagnostic Biomarkers
Tissue-Based Samples
Liquid Biopsies
4.2.3. LncRNAs as Prognostic Biomarkers
Tissue-Based Samples
Liquid Biopsies
4.2.4. LncRNAs as Predictive Biomarkers of Response to Therapy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maher, E.R. Genomics and epigenomics of renal cell carcinoma. Semin. Cancer Biol. 2013, 23, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Patard, J.-J.; Leray, E.; Rodriguez, A.; Rioux-Leclercq, N.; Guillé, F.; Lobel, B. Correlation between symptom graduation, tumor characteristics and survival in renal cell carcinoma. Eur. Urol. 2003, 44, 226–232. [Google Scholar] [CrossRef]
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef]
- Rosenberg, S.A. Interleukin 2 for patients with renal cancer. Nat. Clin. Pract. Oncol. 2007, 4, 497. [Google Scholar] [CrossRef]
- Ikarashi, D.; Nakamura, Y.; Shimodate, H.; Usui, Y.; Ujiie, T.; Obara, W. Complete response to perioperative treatment using nivolumab for metastatic renal cell carcinoma: A case report. Urol. Case Rep. 2019, 24, 100839. [Google Scholar] [CrossRef]
- Wood, C.; Srivastava, P.; Bukowski, R.; Lacombe, L.; Gorelov, A.I.; Gorelov, S.; Mulders, P.; Zieliński, H.; Hoos, A.; Teofilovici, F.; et al. An adjuvant autologous therapeutic vaccine (HSPPC-96; vitespen) versus observation alone for patients at high risk of recurrence after nephrectomy for renal cell carcinoma: A multicentre, open-label, randomised phase III trial. Lancet 2008, 372, 145–154. [Google Scholar] [CrossRef]
- Janowitz, T.; Welsh, S.J.; Zaki, K.; Mulders, P.; Eisen, T. Adjuvant Therapy in Renal Cell Carcinoma—Past, Present, and Future. Semin. Oncol. 2013, 40, 482–491. [Google Scholar] [CrossRef] [Green Version]
- Chamie, K.; Donin, N.M.; Klöpfer, P.; Bevan, P.; Fall, B.; Wilhelm, O.; Störkel, S.; Said, J.; Gambla, M.; Hawkins, R.E.; et al. Adjuvant Weekly Girentuximab Following Nephrectomy for High-Risk Renal Cell Carcinoma. JAMA Oncol. 2017, 3, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Haas, N.B.; Manola, J.; Dutcher, J.P.; Flaherty, K.T.; Uzzo, R.G.; Atkins, M.B.; DiPaola, R.S.; Choueiri, T.K. Adjuvant Treatment for High-Risk Clear Cell Renal Cancer: Updated Results of a High-Risk Subset of the ASSURE Randomized Trial. JAMA Oncol. 2017, 3, 1249–1252. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Haas, N.B.; Donskov, F.; Gross-Goupil, M.; Varlamov, S.; Kopyltsov, E.; Lee, J.L.; Melichar, B.; Rini, B.I.; Choueiri, T.K.; et al. Randomized Phase III Trial of Adjuvant Pazopanib Versus Placebo After Nephrectomy in Patients With Localized or Locally Advanced Renal Cell Carcinoma. J. Clin. Oncol. 2017, 35, 3916–3923. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Ravaud, A.; Patard, J.-J.; Pandha, H.S.; George, D.J.; Patel, A.; Chang, Y.-H.; Escudier, B.; Donskov, F.; Magheli, A.; et al. Adjuvant Sunitinib for High-risk Renal Cell Carcinoma after Nephrectomy: Subgroup Analyses and Updated Overall Survival Results. Eur. Urol. 2017, 73, 62–68. [Google Scholar] [CrossRef]
- Patard, J.-J.; Pignot, G.; Escudier, B.; Eisen, T.; Bex, A.; Sternberg, C.; Rini, B.; Roigas, J.; Choueiri, T.; Bukowski, R.; et al. ICUD-EAU International Consultation on Kidney Cancer 2010: Treatment of Metastatic Disease. Eur. Urol. 2011, 60, 684–690. [Google Scholar] [CrossRef]
- Motzer, R.J.; Hutson, T.E.; Tomczak, P.; Michaelson, M.D.; Bukowski, R.M.; Rixe, O.; Oudard, S.; Négrier, S.; Szczylik, C.; Kim, S.T.; et al. Sunitinib versus Interferon Alfa in Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 2007, 356, 115–124. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Escudier, B.; Powles, T.; Mainwaring, P.N.; Rini, B.I.; Donskov, F.; Hammers, H.; Hutson, T.E.; Lee, J.-L.; Peltola, K.; et al. Cabozantinib versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1814–1823. [Google Scholar] [CrossRef]
- Ratta, R.; Verzoni, E.; Di Maio, M.; Grassi, P.; Colecchia, M.; Fucà, G.; De Braud, F.; Procopio, G. Exposure to Multiple Lines of Treatment and Survival of Patients With Metastatic Renal Cell Carcinoma: A Real-world Analysis. Clin. Genitourin. Cancer 2018, 16, e735–e742. [Google Scholar] [CrossRef]
- Battelli, C.; Cho, D.C. mTOR inhibitors in renal cell carcinoma. Therapy 2011, 8, 359–367. [Google Scholar] [CrossRef] [Green Version]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef]
- McDermott, D.F.; Sosman, J.A.; Sznol, M.; Massard, C.; Gordon, M.S.; Hamid, O.; Powderly, J.D.; Infante, J.R.; Fassò, M.; Wang, Y.V.; et al. Atezolizumab, an Anti–Programmed Death-Ligand 1 Antibody, in Metastatic Renal Cell Carcinoma: Long-Term Safety, Clinical Activity, and Immune Correlates From a Phase Ia Study. J. Clin. Oncol. 2016, 34, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Flaherty, K. Clinical effect and future considerations for molecularly-targeted therapy in renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2008, 26, 543–549. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in Cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Henrique, R.; Luís, A.S.; Jerónimo, C. The Epigenetics of Renal Cell Tumors: From Biology to Biomarkers. Front. Genet. 2012, 3, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, M.R.; Latif, F. The epigenetic landscape of renal cancer. Nat. Rev. Nephrol. 2016, 13, 47–60. [Google Scholar] [CrossRef]
- Jerónimo, C.; Henrique, R. Epigenetic biomarkers in urological tumors: A systematic review. Cancer Lett. 2014, 342, 264–274. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [Green Version]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Boil. 2013, 14, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Boil. 2014, 25, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Boil. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2010, 12, 19–31. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.W. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2015, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, A.; Zarone, M.R.; Irace, C.; Zappavigna, S.; Lombardi, A.; Kawasaki, H.; Caraglia, M.; Misso, G. Non-coding RNAs as a new dawn in tumor diagnosis. Semin. Cell Dev. Boil. 2018, 78, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Publisher Correction: Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Boil. 2019, 20, 321. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Boil. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef] [Green Version]
- Vagin, V.V.; Sigova, A.; Li, C.; Gvozdev, V.; Seitz, H.; Zamore, P.D. A Distinct Small RNA Pathway Silences Selfish Genetic Elements in the Germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Sachidanandam, R.; Hannon, G.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2018, 20, 89–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down–regulation of micro– RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Research Network; Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [CrossRef] [PubMed] [Green Version]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Guil, S.; Esteller, M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int. J. Biochem. Cell Biol. 2009, 41, 87–95. [Google Scholar] [CrossRef]
- Takamizawa, J.; Chamoto, K.; Tsuji, T.; Funamoto, H.; Kosaka, A.; Matsuzaki, J.; Sato, T.; Konishi, H.; Fujio, K.; Yamamoto, K.; et al. Reduced Expression of the let-7 MicroRNAs in Human Lung Cancers in Association with Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, G.; Hayashi, K.; Xi, Y.; Kudo, K.; Uchida, K.; Takasaki, K.; Yamamoto, M.; Ju, J. Non–coding MicroRNAs hsa–let–7g and hsa–miR–181b are Associated with Chemoresponse to S–1 in Colon Cancer. Cancer Genom. Proteom. 2006, 3, 317–324. [Google Scholar]
- Cheng, Y.; Wang, Q.; Jiang, W.; Bian, Y.; Zhou, Y.; Gou, A.; Zhang, W.; Fu, K.; Shi, W. Emerging roles of piRNAs in cancer: Challenges and prospects. Aging 2019, 11, 9932–9946. [Google Scholar] [CrossRef]
- Yu, Y.; Xiao, J.; Hann, S.S. The emerging roles of PIWI-interacting RNA in human cancers. Cancer Manag. Res. 2019, 11, 5895–5909. [Google Scholar] [CrossRef] [Green Version]
- Busch, J.; Ralla, B.; Jung, M.; Wotschofsky, Z.; Trujillo-Arribas, E.; Schwabe, P.; Kilic, E.; Fendler, A.; Jung, K. Piwi–interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J. Exp. Clin. Cancer Res. 2015, 34, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wu, X.; Gao, H.; Jin, J.M.; Li, A.X.; Kim, Y.S.; Pal, S.K.; Nelson, R.A.; Lau, C.M.; Guo, C.; et al. Piwi-Interacting RNAs (piRNAs) Are Dysregulated in Renal Cell Carcinoma and Associated with Tumor Metastasis and Cancer-Specific Survival. Mol. Med. 2015, 21, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Iliev, R.; Fedorko, M.; Machackova, T.; Mlcochova, H.; Svoboda, M.; Pacik, D.; Dolezel, J.; Stanik, M.; Capoor, M.N. Expression Levels of PIWI-interacting RNA, piR-823, Are Deregulated in Tumor Tissue, Blood Serum and Urine of Patients with Renal Cell Carcinoma. Anticancer. Res. 2016, 36, 6419–6424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wotschofsky, Z.; Busch, J.; Jung, M.; Kempkensteffen, C.; Weikert, S.; Schaser, K.D.; Melcher, I.; Kilic, E.; Miller, K.; Kristiansen, G.; et al. Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clin. Chim. Acta 2013, 416, 5–10. [Google Scholar] [CrossRef]
- Silva-Santos, R.M.; Costa-Pinheiro, P.; Luis, A.; Antunes, L.; Lobo, F.; Oliveira, J.; Henrique, R.; Jerónimo, C. MicroRNA profile: A promising ancillary tool for accurate renal cell tumour diagnosis. Br. J. Cancer 2013, 109, 2646–2653. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Mollenkopf, H.-J.; Grimm, C.H.; Wagner, I.; Albrecht, M.; Waller, T.; Pilarsky, C.; Johannsen, M.; Stephan, C.; Lehrach, H.; et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J. Cell. Mol. Med. 2009, 13, 3918–3928. [Google Scholar] [CrossRef] [Green Version]
- Fridman, E.; Dotan, Z.; Barshack, I.; Ben David, M.; Dov, A.; Tabak, S.; Zion, O.; Benjamin, S.; Benjamin, H.; Kuker, H.; et al. Accurate Molecular Classification of Renal Tumors Using MicroRNA Expression. J. Mol. Diagn. 2010, 12, 687–696. [Google Scholar] [CrossRef]
- Faragalla, H.; Youssef, Y.M.; Scorilas, A.; Khalil, B.; White, N.M.; Mejia-Guerrero, S.; Khella, H.; Jewett, M.A.; Evans, A.; Lichner, Z.; et al. The Clinical Utility of miR-21 as a Diagnostic and Prognostic Marker for Renal Cell Carcinoma. J. Mol. Diagn. 2012, 14, 385–392. [Google Scholar] [CrossRef]
- Wach, S.; Nolte, E.; Theil, A.; Stöhr, C.; Rau, T.T.; Hartmann, A.; Ekici, A.; Keck, B.; Taubert, H.; Wullich, B. MicroRNA profiles classify papillary renal cell carcinoma subtypes. Br. J. Cancer 2013, 109, 714–722. [Google Scholar] [CrossRef] [Green Version]
- Zaravinos, A.; Lambrou, G.I.; Mourmouras, N.; Katafygiotis, P.; Papagregoriou, G.; Giannikou, K.; Delakas, D.; Deltas, C. New miRNA Profiles Accurately Distinguish Renal Cell Carcinomas and Upper Tract Urothelial Carcinomas from the Normal Kidney. PLoS ONE 2014, 9, e91646. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Ruan, A.; Han, W.; Zhao, Y.; Lu, X.; Xiao, P.; Shi, H.; Wang, R.; Chen, L.; et al. miR-141 Is a Key Regulator of Renal Cell Carcinoma Proliferation and Metastasis by Controlling EphA2 Expression. Clin. Cancer Res. 2014, 20, 2617–2630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ruan, A.; Wang, X.; Han, W.; Wang, R.; Lou, N.; Ruan, H.; Qiu, B.; Yang, H.; Zhang, X. miR–129–3p, as a diagnostic and prognostic biomarker for renal cell carcinoma, attenuates cell migration and invasion via downregulating multiple metastasis–related genes. J. Cancer Res. Clin. Oncol. 2014, 140, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Vergho, D.; Kneitz, S.; Kalogirou, C.; Burger, M.; Krebs, M.; Rosenwald, A.; Spahn, M.; Löser, A.; Kocot, A.; Riedmiller, H.; et al. Impact of miR-21, miR-126 and miR-221 as Prognostic Factors of Clear Cell Renal Cell Carcinoma with Tumor Thrombus of the Inferior Vena Cava. PLoS ONE 2014, 9, e109877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulos, E.I.; Petraki, C.; Gregorakis, A.; Fragoulis, E.G.; Scorilas, A. Clinical evaluation of microRNA-145 expression in renal cell carcinoma: A promising molecular marker for discriminating and staging the clear cell histological subtype. Boil. Chem. 2016, 397, 529–539. [Google Scholar] [CrossRef]
- Liep, J.; Kilic, E.; Meyer, H.A.; Busch, J.; Jung, K.; Rabien, A. Cooperative Effect of miR–141–3p and miR–145–5p in the Regulation of Targets in Clear Cell Renal Cell Carcinoma. PLoS ONE 2016, 11, e0157801. [Google Scholar] [CrossRef] [PubMed]
- Kowalik, C.G.; Palmer, D.A.; Sullivan, T.B.; Teebagy, P.A.; Dugan, J.M.; Libertino, J.A.; Burks, E.J.; Canes, D.; Rieger-Christ, K.M. Profiling microRNA from nephrectomy and biopsy specimens: Predictors of progression and survival in clear cell renal cell carcinoma. BJU Int. 2017, 120, 428–440. [Google Scholar] [CrossRef] [Green Version]
- Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Hussein, M.H.; Al-Qahtani, S.A.M.; Shaalan, A.A.M. MicroRNA-34a: A Key Regulator in the Hallmarks of Renal Cell Carcinoma. Oxidative Med. Cell. Longev. 2017, 2017, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Di Meo, A.; Saleeb, R.; Wala, S.J.; Khella, H.W.; Ding, Q.; Zhai, H.; Krishan, K.; Krizova, A.; Gabril, M.; Evans, A.; et al. A miRNA-based classification of renal cell carcinoma subtypes by PCR and in situ hybridization. Oncotarget 2017, 9, 2092–2104. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.; Khandelwal, M.; Seth, A.; Saini, A.; Dogra, P.N.; Sharma, A. Serum microRNA Expression Profiling: Potential Diagnostic Implications of a Panel of Serum microRNAs for Clear Cell Renal Cell Cancer. Urology 2017, 104, 64–69. [Google Scholar] [CrossRef]
- Bhat, N.S.; Colden, M.; Dar, A.A.; Saini, S.; Arora, P.; Shahryari, V.; Yamamura, S.; Tanaka, Y.; Kato, T.; Majid, S.; et al. MicroRNA–720 Regulates E–cadherin–alphaE–catenin Complex and Promotes Renal Cell Carcinoma. Mol. Cancer Ther. 2017, 16, 2840–2848. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, P.; Kulkarni, P.; Majid, S.; Shahryari, V.; Hashimoto, Y.; Bhat, N.S.; Shiina, M.; Deng, G.; Saini, S.; Tabatabai, Z.L.; et al. MicroRNA-203 Inhibits Long Noncoding RNA HOTAIR and Regulates Tumorigenesis through Epithelial-to-mesenchymal Transition Pathway in Renal Cell Carcinoma. Mol. Cancer Ther. 2018, 17, 1061–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, P.; Dasgupta, P.; Bhat, N.S.; Shahryari, V.; Shiina, M.; Hashimoto, Y.; Majid, S.; Deng, G.; Saini, S.; Tabatabai, Z.L.; et al. Elevated miR–182–5p Associates with Renal Cancer Cell Mitotic Arrest through Diminished MALAT–1 Expression. Mol. Cancer Res. 2018, 16, 1750–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Lou, N.; Ruan, A.; Qiu, B.; Yan, Y.; Wang, X.; Du, Q.; Ruan, H.; Han, W.; Wei, H.; et al. miR–224/miR–141 ratio as a novel diagnostic biomarker in renal cell carcinoma. Oncol. Lett. 2018, 16, 1666–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, W.; Li, S.; Zhang, J.; Chen, Y.; Ma, J.; Kong, W.; Gong, D.; Zheng, J.; Xue, W.; Xu, Y. Sunitinib–suppressed miR–452–5p facilitates renal cancer cell invasion and metastasis through modulating SMAD4/SMAD7 signals. Mol. Cancer 2018, 17, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Tolkach, Y.; Schmidt, D.; Toma, M.; Muders, M.H.; Kristiansen, G.; Müller, S.C.; Ellinger, J. Mitochondrial PIWI-interacting RNAs are novel biomarkers for clear cell renal cell carcinoma. World J. Urol. 2018, 37, 1639–1647. [Google Scholar] [CrossRef]
- Li, J.; Chen, C.; Shi, Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J. Cell. Physiol. 2017, 233, 4458–4465. [Google Scholar] [CrossRef]
- Wulfken, L.M.; Moritz, R.; Ohlmann, C.; Holdenrieder, S.; Jung, V.; Becker, F.; Herrmann, E.; Walgenbach-Brünagel, G.; Von Ruecker, A.; Müller, S.C.; et al. MicroRNAs in Renal Cell Carcinoma: Diagnostic Implications of Serum miR-1233 Levels. PLoS ONE 2011, 6, e25787. [Google Scholar] [CrossRef] [Green Version]
- Zhao, A.; Li, G.; Péoc’H, M.; Genin, C.; Gigante, M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp. Mol. Pathol. 2013, 94, 115–120. [Google Scholar] [CrossRef]
- Zhang, W.; Ni, M.; Su, Y.; Wang, H.; Zhu, S.; Zhao, A.; Li, G. MicroRNAs in Serum Exosomes as Potential Biomarkers in Clear-cell Renal Cell Carcinoma. Eur. Urol. Focus 2018, 4, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Tusong, H.; Maolakuerban, N.; Guan, J.; Rexiati, M.; Wang, W.-G.; Azhati, B.; Nuerrula, Y.; Wang, Y.-J. Functional analysis of serum microRNAs miR-21 and miR-106a in renal cell carcinoma. Cancer Biomark. 2017, 18, 79–85. [Google Scholar] [CrossRef]
- Lou, N.; Ruan, A.-M.; Qiu, B.; Bao, L.; Xu, Y.-C.; Zhao, Y.; Sun, R.-L.; Zhang, S.-T.; Xu, G.-H.; Ruan, H.-L.; et al. miR–144–3p as a novel plasma diagnostic biomarker for clear cell renal cell carcinoma. Urol. Oncol. 2017, 35, 36.e7. [Google Scholar] [CrossRef] [PubMed]
- Butz, H.; Nofech-Mozes, R.; Ding, Q.; Khella, H.W.; Szabó, P.M.; Jewett, M.; Finelli, A.; Lee, J.; Ordon, M.; Stewart, R.; et al. Exosomal MicroRNAs Are Diagnostic Biomarkers and Can Mediate Cell–Cell Communication in Renal Cell Carcinoma. Eur. Urol. Focus 2016, 2, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Mytsyk, Y.; Dosenko, V.; Borys, Y.; Kucher, A.; Gazdikova, K.; Büsselberg, D.; Caprnda, M.; Kruzliak, P.; Farooqi, A.A.; Lubov, M. MicroRNA-15a expression measured in urine samples as a potential biomarker of renal cell carcinoma. Int. Urol. Nephrol. 2018, 50, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Redova, M.; Poprach, A.; Nekvindova, J.; Iliev, R.; Radova, L.; Lakomy, R.; Svoboda, M.; Vyzula, R.; Slaby, O. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J. Transl. Med. 2012, 10, 55. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, H.; Kanda, Y.; Sejima, T.; Osaki, M.; Okada, F.; Takenaka, A. Serum miR-210 as a potential biomarker of early clear cell renal cell carcinoma. Int. J. Oncol. 2013, 44, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, A.L.; Ferreira, M.; Silva, J.; Gomes, M.; Dias, F.; Santos, J.I.; Maurício, J.; Lobo, F.; Medeiros, R. Higher circulating expression levels of miR-221 associated with poor overall survival in renal cell carcinoma patients. Tumor Boil. 2013, 35, 4057–4066. [Google Scholar] [CrossRef]
- Wang, C.; Hu, J.; Lu, M.; Gu, H.; Zhou, X.; Chen, X.; Zen, K.; Zhang, C.-Y.; Zhang, T.; Ge, J.; et al. A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci. Rep. 2015, 5, 7610. [Google Scholar] [CrossRef]
- Fedorko, M.; Stanik, M.; Iliev, R.; Rédová-Lojová, M.; Machackova, T.; Svoboda, M.; Pacík, D.; Dolezel, J.; Slaby, O. Combination of MiR-378 and MiR-210 Serum Levels Enables Sensitive Detection of Renal Cell Carcinoma. Int. J. Mol. Sci. 2015, 16, 23382–23389. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhao, A.; Péoch, M.; Cottier, M.; Mottet, N. Detection of urinary cell-free miR-210 as a potential tool of liquid biopsy for clear cell renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2017, 35, 294–299. [Google Scholar] [CrossRef]
- Fedorko, M.; Juracek, J.; Stanik, M.; Svoboda, M.; Poprach, A.; Buchler, T.; Pacik, D.; Dolezel, J.; Slaby, O. Detection of let-7 miRNAs in urine supernatant as potential diagnostic approach in non-metastatic clear-cell renal cell carcinoma. Biochem. Med. 2017, 27, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Chanudet, E.; Wozniak, M.B.; Bouaoun, L.; Byrnes, G.B.; Mukeria, A.; Zaridze, D.G.; Brennan, P.; Muller, D.C.; Scelo, G. Large-scale genome-wide screening of circulating microRNAs in clear cell renal cell carcinoma reveals specific signatures in late-stage disease. Int. J. Cancer 2017, 141, 1730–1740. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, T.; Chen, C.; Wu, Z.; Bai, P.; Li, S.; Chen, B.; Liu, R.; Zhang, K.; Li, W.; et al. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J. Cell. Biochem. 2018, 120, 1492–1502. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Deng, X.; Zhang, J. Identification of dysregulated serum miR–508–3p and miR–885–5p as potential diagnostic biomarkers of clear cell renal carcinoma. Mol. Med. Rep. 2019, 20, 5075–5083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Meo, A.; Brown, M.D.; Finelli, A.; Jewett, M.A.; Diamandis, E.P.; Mac-Way, F. Prognostic urinary miRNAs for the assessment of small renal masses. Clin. Biochem. 2019, 75, 15–22. [Google Scholar] [CrossRef]
- Outeiro–Pinho, G.; Barros-Silva, D.; Aznar, E.; Sousa, A.-I.; Vieira-Coimbra, M.; Oliveira, J.; Gonçalves, C.S.; Costa, B.M.; Junker, K.; Henrique, R.; et al. MicroRNA–30a–5p(me): A novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma in tissue and urine samples. J. Exp. Clin. Cancer Res. 2020, 39, 98. [Google Scholar] [CrossRef]
- Wang, G.; Chen, L.; Meng, J.; Chen, M.; Zhuang, L.; Zhang, L. Overexpression of microRNA-100 predicts an unfavorable prognosis in renal cell carcinoma. Int. Urol. Nephrol. 2013, 45, 373–379. [Google Scholar] [CrossRef]
- Zhao, J.-J.; Chen, P.-J.; Duan, R.-Q.; Li, K.-J.; Wang, Y.-Z.; Li, Y. Up-regulation of miR-630 in clear cell renal cell carcinoma is associated with lower overall survival. Int. J. Clin. Exp. Pathol. 2014, 7, 3318–3323. [Google Scholar]
- Samaan, S.; Khella, H.W.; Girgis, A.; Scorilas, A.; Lianidou, E.; Gabril, M.; Krylov, S.N.; Jewett, M.; Bjarnason, G.A.; El-Said, H.; et al. miR-210 Is a Prognostic Marker in Clear Cell Renal Cell Carcinoma. J. Mol. Diagn. 2015, 17, 136–144. [Google Scholar] [CrossRef]
- Dias, F.; Teixeira, A.; Ferreira, M.; Adem, B.; Bastos, N.; Vieira, J.; Fernandes, M.; Sequeira, M.I.; Maurício, J.; Lobo, F.; et al. Plasmatic miR-210, miR-221 and miR-1233 profile: Potential liquid biopsies candidates for renal cell carcinoma. Oncotarget 2017, 8, 103315–103326. [Google Scholar] [CrossRef]
- Zhang, J.; Ye, Y.; Chang, D.W.; Lin, S.-H.; Huang, M.; Tannir, N.M.; Matin, S.; Karam, J.A.; Wood, C.G.; Chen, Z.-N.; et al. Global and Targeted miRNA Expression Profiling in Clear Cell Renal Cell Carcinoma Tissues Potentially Links miR–155–5p and miR–210–3p to both Tumorigenesis and Recurrence. Am. J. Pathol. 2018, 188, 2487–2496. [Google Scholar] [CrossRef] [Green Version]
- McCormick, R.I.; Blick, C.; Ragoussis, J.; Schoedel, J.; Mole, D.R.; Young, A.C.; Selby, P.J.; Banks, R.; Harris, A.L. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br. J. Cancer 2013, 108, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Nakata, W.; Uemura, M.; Sato, M.; Fujita, K.; Jingushi, K.; Ueda, Y.; Kitae, K.; Tsujikawa, K.; Nonomura, N. Expression of miR-27a-3p is an independent predictive factor for recurrence in clear cell renal cell carcinoma. Oncotarget 2015, 6, 21645–21654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinmei, S.; Sakamoto, N.; Goto, K.; Sentani, K.; Anami, K.; Hayashi, T.; Teishima, J.; Matsubara, A.; Oue, N.; Kitadai, Y.; et al. MicroRNA-155 is a predictive marker for survival in patients with clear cell renal cell carcinoma. Int. J. Urol. 2012, 20, 468–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrandt, M.A.T.; Gu, J.; Lin, J.; Ye, Y.; Tan, W.; Tamboli, P.; Wood, C.G.; Wu, X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 2010, 29, 5724–5728. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Weng, L.; Li, X.; Guo, C.; Pal, S.K.; Jin, J.M.; Li, Y.; Nelson, R.A.; Mu, B.; Onami, S.H.; et al. Identification of a 4-microRNA Signature for Clear Cell Renal Cell Carcinoma Metastasis and Prognosis. PLoS ONE 2012, 7, e35661. [Google Scholar] [CrossRef]
- Gebauer, K.; Peters, I.; Dubrowinskaja, N.; Hennenlotter, J.; Abbas, M.; Scherer, R.; Tezval, H.; Merseburger, A.S.; Stenzl, A.; A Kuczyk, M.; et al. Hsa–mir–124–3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br. J. Cancer 2013, 108, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Goto, K.; Oue, N.; Shinmei, S.; Sentani, K.; Sakamoto, N.; Naito, Y.; Hayashi, T.; Teishima, J.; Matsubara, A.; Yasui, W. Expression of miR-486 is a potential prognostic factor after nephrectomy in advanced renal cell carcinoma. Mol. Clin. Oncol. 2012, 1, 235–240. [Google Scholar] [CrossRef]
- Vergho, D.C.; Kneitz, S.; Rosenwald, A.; Scherer, C.; Spahn, M.; Burger, M.; Riedmiller, H.; Kneitz, B. Combination of expression levels of miR-21 and miR-126 is associated with cancer-specific survival in clear-cell renal cell carcinoma. BMC Cancer 2014, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Fritz, H.K.; Lindgren, D.; Ljungberg, B.; Axelson, H.; Dahlbäck, B. The miR(21/10b) ratio as a prognostic marker in clear cell renal cell carcinoma. Eur. J. Cancer 2014, 50, 1758–1765. [Google Scholar] [CrossRef] [Green Version]
- Fu, Q.; Liu, Z.; Pan, D.; Zhang, W.; Xu, L.; Zhu, Y.; Liu, H.; Xu, J. Tumor miR-125b predicts recurrence and survival of patients with clear-cell renal cell carcinoma after surgical resection. Cancer Sci. 2014, 105, 1427–1434. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Zhao, Z.; Xu, W.; Hou, J.; Du, X. Down-regulation of miR-497 is associated with poor prognosis in renal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 758–764. [Google Scholar] [PubMed]
- Khella, H.W.; Scorilas, A.; Mozes, R.; Mirham, L.; Lianidou, E.; Krylov, S.N.; Lee, J.Y.; Ordon, M.; Stewart, R.; Jewett, M.A.; et al. Low Expression of miR-126 Is a Prognostic Marker for Metastatic Clear Cell Renal Cell Carcinoma. Am. J. Pathol. 2015, 185, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.-Q.; Zhang, H.-M.; Chen, S.-J.; Yan, Y.; Zheng, J.-H. MiR–506 is down–regulated in clear cell renal cell carcinoma and inhibits cell growth and metastasis via targeting FLOT. PLoS ONE 2015, 10, e0120258. [Google Scholar]
- Xu, M.; Gu, M.; Zhang, K.; Zhou, J.; Wang, Z.; Da, J. miR-203 inhibition of renal cancer cell proliferation, migration and invasion by targeting of FGF. Diagn. Pathol. 2015, 10, 24. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Xu, H. Prognostic value of meta-signature miRNAs in renal cell carcinoma: An integrated miRNA expression profiling analysis. Sci. Rep. 2015, 5, 10272. [Google Scholar] [CrossRef] [Green Version]
- Butz, H.; Szabó, P.M.; Khella, H.W.; Nofech-Mozes, R.; Patócs, A.; Mac-Way, F. miRNA-target network reveals miR-124as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT. Oncotarget 2015, 6, 12543–12557. [Google Scholar] [CrossRef] [Green Version]
- Nofech-Mozes, R.; Khella, H.W.Z.; Scorilas, A.; Youssef, L.; Krylov, S.N.; Lianidou, E.; Sidiropoulos, K.G.; Gabril, M.; Evans, A.; Mac-Way, F. MicroRNA-194 is a Marker for Good Prognosis in Clear Cell Renal Cell Carcinoma. Cancer Med. 2016, 5, 656–664. [Google Scholar] [CrossRef]
- Ma, Q.; Peng, Z.; Wang, L.; Li, Y.; Wang, K.; Zheng, J.; Liang, Z.; Liu, T. miR-19a correlates with poor prognosis of clear cell renal cell carcinoma patients via promoting cell proliferation and suppressing PTEN/SMAD4 expression. Int. J. Oncol. 2016, 49, 2589–2599. [Google Scholar] [CrossRef] [Green Version]
- García-Donás, J.; Beuselinck, B.; Inglada-Pérez, L.; Castro, O.G.; Schöffski, P.; Wozniak, A.; Bechter, O.; Apellániz-Ruiz, M.; Leandro-García, L.J.; Esteban, E.; et al. Deep sequencing reveals microRNAs predictive of antiangiogenic drug response. JCI Insight 2016, 1, e86051. [Google Scholar] [CrossRef] [Green Version]
- Khella, H.W.Z.; Daniel, N.; Youssef, L.; Scorilas, A.; Nofech-Mozes, R.; Mirham, L.; Krylov, S.N.; Liandeau, E.; Krizova, A.; Finelli, A.; et al. miR-10b is a prognostic marker in clear cell renal cell carcinoma. J. Clin. Pathol. 2017, 70, 854–859. [Google Scholar] [CrossRef]
- Chen, C.; Xue, S.; Zhang, J.; Chen, W.; Gong, D.; Zheng, J.; Ma, J.; Xue, W.; Chen, Y.; Zhai, W.; et al. DNA–methylation–mediated repression of miR–766–3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int. J. Cancer 2017, 141, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Quan, J.; Li, Z.; Zhao, L.; Zhou, L.; Jinling, X.; Weijie, X.; Guan, X.; Li, H.; Yang, S.; et al. miR-566 functions as an oncogene and a potential biomarker for prognosis in renal cell carcinoma. Biomed. Pharmacother. 2018, 102, 718–727. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, Z.; Pan, X.; Lai, Y.; Quan, J.; Zhao, L.; Xu, J.; Xu, W.; Guan, X.; Li, H.; et al. Identification of miR-18a-5p as an oncogene and prognostic biomarker in RCC. Am. J. Transl. Res. 2018, 10, 1874–1886. [Google Scholar] [PubMed]
- Zhou, L.; Pan, X.; Li, Z.; Chen, P.; Quan, J.; Lin, C.; Lai, Y.; Xu, J.; Xu, W.; Guan, X.; et al. Oncogenic miR-663a is associated with cellular function and poor prognosis in renal cell carcinoma. Biomed. Pharmacother. 2018, 105, 1155–1163. [Google Scholar] [CrossRef]
- Pan, X.; Li, Z.; Zhao, L.; Quan, J.; Zhou, L.; Xu, J.; Xu, W.; Guan, X.; Li, H.; Yang, S.; et al. microRNA-572 functions as an oncogene and a potential biomarker for renal cell carcinoma prognosis. Oncol. Rep. 2018, 40, 3092–3101. [Google Scholar] [CrossRef] [Green Version]
- Quan, J.; Pan, X.; Li, Y.; Hu, Y.; Tao, L.; Li, Z.; Zhao, L.; Wang, J.; Li, H.; Lai, Y.; et al. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed. Pharmacother. 2019, 110, 656–666. [Google Scholar] [CrossRef]
- Li, H.; Pan, X.; Gui, Y.; Quan, J.; Li, Z.; Zhao, L.; Guan, X.; Xu, J.; Xu, W.; Lai, Y. Upregulation of miR–183–5p predicts worse survival in patients with renal cell cancer after surgery. Cancer Biomark. 2019, 24, 153–158. [Google Scholar] [CrossRef]
- Chen, X. Expression of microRNA-3133 correlates with the prognosis in patients with clear cell renal cell carcinoma. Medicine 2019, 98, e16008. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Y.; Li, W.; Yu, S.; Wen, Z.; Chen, Z.; Lin, F. miR–221–5p acts as an oncogene and predicts worse survival in patients of renal cell cancer. Biomed. Pharmacother. 2019, 119, 109406. [Google Scholar] [CrossRef]
- Peng, X.; Pan, X.; Liu, K.; Zhang, C.; Zhao, L.; Li, H.; Guan, X.; Xu, W.; Xu, J.; Zhang, F.; et al. miR–142–3p as a novel biomarker for predicting poor prognosis in renal cell carcinoma patients after surgery. Int. J. Biol. Markers 2019, 34, 302–308. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Pan, X.; Peng, X.; Zhang, C.; Li, H.; Guan, X.; Xu, W.; Xu, J.; Zhao, L.; Wang, T.; et al. Associations of high expression of miR-106b-5p detected from FFPE sample with poor prognosis of RCC patients. Pathol. Res. Pract. 2019, 215, 152391. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann, J.; Arndt, M.; Pleyers, R.; Fehlmann, T.; Hoelters, S.; Zeuschner, P.; Vogt, A.; Pryalukhin, A.; Schaeffeler, E.; Bohle, R.M.; et al. 4-miRNA Score Predicts the Individual Metastatic Risk of Renal Cell Carcinoma Patients. Ann. Surg. Oncol. 2019, 26, 3765–3773. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Giridhar, K.V.; Tian, Y.; Tschannen, M.R.; Zhu, J.; Huang, C.-C.; Kilari, D.; Kohli, M.; Wang, L. Plasma exosomal miRNAs-based prognosis in metastatic kidney cancer. Oncotarget 2017, 8, 63703–63714. [Google Scholar] [CrossRef] [Green Version]
- Fujii, N.; Hirata, H.; Ueno, K.; Mori, J.; Oka, S.; Shimizu, K.; Kawai, Y.; Inoue, R.; Yamamoto, Y.; Matsumoto, H.; et al. Extracellular miR-224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget 2017, 8, 109877–109888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinemann, F.G.; Tolkach, Y.; Deng, M.; Schmidt, D.; Perner, S.; Kristiansen, G.; Müller, S.; Ellinger, J. Serum miR–122–5p and miR–206 expression: Non–invasive prognostic biomarkers for renal cell carcinoma. Clin. Epigenetics 2018, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Schwandt, A.; Wood, L.S.; Rini, B.; Dreicer, R. Management of side effects associated with sunitinib therapy for patients with renal cell carcinoma. OncoTargets Ther. 2009, 2, 51–61. [Google Scholar]
- Eisen, T.; Sternberg, C.N.; Robert, C.; Mulders, P.; Pyle, L.; Zbinden, S.; Izzedine, H.; Escudier, B. Targeted Therapies for Renal Cell Carcinoma: Review of Adverse Event Management Strategies. J. Natl. Cancer Inst. 2012, 104, 93–113. [Google Scholar] [CrossRef] [Green Version]
- Makhov, P.; Joshi, S.; Ghatalia, P.; Kutikov, A.; Uzzo, R.; Kolenko, V.M. Resistance to Systemic Therapies in Clear Cell Renal Cell Carcinoma: Mechanisms and Management Strategies. Mol. Cancer Ther. 2018, 17, 1355–1364. [Google Scholar] [CrossRef] [Green Version]
- Institute, N.C. Precision Medicine in Cancer Treatment. 3 October 2017. Available online: https://www.cancer.gov/about-cancer/treatment/types/precision-medicine (accessed on 1 April 2020).
- Corrà, F.; Agnoletto, C.; Minotti, L.; Baldassari, F.; Volinia, S. The Network of Non-coding RNAs in Cancer Drug Resistance. Front. Oncol. 2018, 8, 327. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, K.; Zhou, H.; Wu, Y.; Li, C.; Liu, Y.; Liu, Z.; Xu, Q.; Liu, S.; Xiao, D.; et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 2020, 19, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berkers, J.; Govaere, O.; Wolter, P.; Beuselinck, B.; Schöffski, P.; Van Kempen, L.C.; Albersen, M.; Oord, J.V.D.; Roskams, T.; Swinnen, J.; et al. A Possible Role for MicroRNA-141 Down-Regulation in Sunitinib Resistant Metastatic Clear Cell Renal Cell Carcinoma Through Induction of Epithelial-to-Mesenchymal Transition and Hypoxia Resistance. J. Urol. 2013, 189, 1930–1938. [Google Scholar] [CrossRef] [PubMed]
- Merhautová, J.; Hezova, R.; Poprach, A.; Kovarikova, A.; Radova, L.; Svoboda, M.; Vyzula, R.; Demlova, R.; Slaby, O. miR-155 and miR-484 Are Associated with Time to Progression in Metastatic Renal Cell Carcinoma Treated with Sunitinib. BioMed Res. Int. 2015, 2015, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Go, H.; Kang, M.J.; Kim, P.-J.; Lee, J.-L.; Park, J.Y.; Park, J.-M.; Ro, J.Y.; Cho, Y.M. Development of Response Classifier for Vascular Endothelial Growth Factor Receptor (VEGFR)-Tyrosine Kinase Inhibitor (TKI) in Metastatic Renal Cell Carcinoma. Pathol. Oncol. Res. 2017, 25, 51–58. [Google Scholar] [CrossRef]
- Gámez-Pozo, A.; Antón-Aparicio, L.M.; Bayona, C.; Borrega, P.; Sancho, M.I.G.; García-Domínguez, R.; De Portugal, T.; Ramos-Vázquez, M.; Pérez-Carrión, R.; Bolós, M.V.; et al. MicroRNA Expression Profiling of Peripheral Blood Samples Predicts Resistance to First-line Sunitinib in Advanced Renal Cell Carcinoma Patients. Neoplasia 2012, 14, 1144–1152. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Duan, L.; Yin, G.; Tan, J.; Jiang, X. miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J. Chemother. 2013, 25, 229–238. [Google Scholar] [CrossRef]
- Prior, C.; Perez-Gracia, J.L.; García-Donás, J.; Rodriguez-Antona, C.; Guruceaga, E.; Esteban, E.; Suárez, C.; Castellano, D.; Del Alba, A.G.; Lozano, M.D.; et al. Identification of Tissue microRNAs Predictive of Sunitinib Activity in Patients with Metastatic Renal Cell Carcinoma. PLoS ONE 2014, 9, e86263. [Google Scholar] [CrossRef]
- Gao, C.; Peng, F.H.; Peng, L.K. MiR-200c sensitizes clear-cell renal cell carcinoma cells to sorafenib and imatinib by targeting heme oxygenase-1. Neoplasma 2014, 61, 680–689. [Google Scholar] [CrossRef] [Green Version]
- Mu, W.; Hu, C.; Zhang, H.; Qu, Z.; Cen, J.; Qiu, Z.; Li, C.; Ren, H.; Li, Y.; He, X.; et al. miR-27b synergizes with anticancer drugs via p53 activation and CYP1B1 suppression. Cell Res. 2015, 25, 477–495. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.; Zhu, H.; Gu, D.; Pan, X.; Qian, L.; Xue, B.; Yang, D.; Zhou, J.; Shan, Y. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem. Biophys. Res. Commun. 2015, 459, 234–239. [Google Scholar] [CrossRef]
- Chang, I.; Mitsui, Y.; Fukuhara, S.; Gill, A.; Wong, D.K.; Yamamura, S.; Shahryari, V.; Tabatabai, Z.L.; Dahiya, R.; Shin, D.M.; et al. Loss of miR-200c up-regulates CYP1B1 and confers docetaxel resistance in renal cell carcinoma. Oncotarget 2015, 6, 7774–7787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, Q.-Z.; Du, Y.-F.; Liu, X.-G.; Li, X.; He, D.-L. miR-124 represses FZD5 to attenuate P-glycoprotein-mediated chemo-resistance in renal cell carcinoma. Tumor Boil. 2015, 36, 7017–7026. [Google Scholar] [CrossRef] [PubMed]
- Khella, H.W.Z.; Butz, H.; Ding, Q.; Rotondo, F.; Evans, K.R.; Kupchak, P.; Dharsee, M.; Latif, A.; Pasic, M.D.; Lianidou, E.; et al. miR–221/222 Are Involved in Response to Sunitinib Treatment in Metastatic Renal Cell Carcinoma. Mol. Ther. 2015, 23, 1748–1758. [Google Scholar] [CrossRef] [PubMed]
- Lukamowicz-Rajska, M.; Mittmann, C.; Prummer, M.; Zhong, Q.; Bedke, J.; Hennenlotter, J.; Stenzl, A.; Mischo, A.; Bihr, S.; Schmidinger, M.; et al. MiR-99b-5p expression and response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients. Oncotarget 2016, 7, 78433–78447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pili, R.; Liu, G.; Chintala, S.; Verheul, H.; Rehman, S.; Attwood, K.; Lodge, M.A.; Wahl, R.; Martin, J.I.; Miles, K.M.; et al. Combination of the histone deacetylase inhibitor vorinostat with bevacizumab in patients with clear-cell renal cell carcinoma: A multicentre, single-arm phase I/II clinical trial. Br. J. Cancer 2017, 116, 874–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puente, J.; Laínez, N.; Dueñas, M.; Méndez-Vidal, M.J.; Esteban, E.; Castellano, D.; Martinez-Fernández, M.; Basterretxea, L.; Juan-Fita, M.J.; Antón, L.; et al. Novel potential predictive markers of sunitinib outcomes in long–term responders versus primary refractory patients with metastatic clear–cell renal cell carcinoma. Oncotarget 2017, 8, 30410–30421. [Google Scholar] [CrossRef]
- Xiao, W.; Lou, N.; Ruan, H.; Bao, L.; Xiong, Z.; Yuan, C.; Tong, J.; Xu, G.; Zhou, Y.; Qu, Y.; et al. Mir-144-3p Promotes Cell Proliferation, Metastasis, Sunitinib Resistance in Clear Cell Renal Cell Carcinoma by Downregulating ARID1A. Cell. Physiol. Biochem. 2017, 43, 2420–2433. [Google Scholar] [CrossRef]
- Sun, X.; Lou, L.; Zhong, K.; Wan, L. MicroRNA-451 regulates chemoresistance in renal cell carcinoma by targeting ATF-2 gene. Exp. Boil. Med. 2017, 242, 1299–1305. [Google Scholar] [CrossRef] [Green Version]
- Kovacova, J.; Juracek, J.; Poprach, A.; Büchler, T.; Kopecký, J.; Fiala, O.; Svoboda, M.; Capoor, M.N. Candidate MicroRNA Biomarkers of Therapeutic Response to Sunitinib in Metastatic Renal Cell Carcinoma: A Validation Study in Patients with Extremely Good and Poor Response. Anticancer Res. 2018, 38, 2961–2965. [Google Scholar] [CrossRef]
- Kovacova, J.; Juracek, J.; Poprach, A.; Kopecky, J.; Fiala, O.; Svoboda, M.; Fabian, P.; Radova, L.; Brabec, P.; Buchler, T.; et al. MiR-376b-3p Is Associated With Long-term Response to Sunitinib in Metastatic Renal Cell Carcinoma Patients. Cancer Genom. Proteom. 2019, 16, 353–359. [Google Scholar] [CrossRef] [Green Version]
- He, J.; He, J.; Min, L.; He, Y.; Guan, H.; Wang, J.; Peng, X. Extracellular vesicles transmitted miR–31–5p promotes sorafenib resistance by targeting MLH1 in renal cell carcinoma. Int. J. Cancer 2020, 146, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Kurozumi, A.; Nohata, N.; Kojima, S.; Matsushita, R.; Yoshino, H.; Yamazaki, K.; Ishida, Y.; Ichikawa, T.; Naya, Y.; et al. The microRNA signature of patients with sunitinib failure: Regulation of UHRF1 pathways by microRNA-101 in renal cell carcinoma. Oncotarget 2016, 7, 59070–59086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prensner, J.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Wang, L.; Ding, Y.; Lu, X.; Zhang, G.; Yang, J.; Zheng, H.; Wang, H.; Jiang, Y.; Xu, L. LncRNA Structural Characteristics in Epigenetic Regulation. Int. J. Mol. Sci. 2017, 18, 2659. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Dammert, M.A.; Grummt, I.; Bierhoff, H. lncRNA-Induced Nucleosome Repositioning Reinforces Transcriptional Repression of rRNA Genes upon Hypotonic Stress. Cell Rep. 2016, 14, 1876–1882. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Ay, F.; Lee, C.; Gülsoy, G.; Deng, X.; Cook, S.; Hesson, J.; Cavanaugh, C.; Ware, C.B.; Krumm, A.; et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat. Methods 2014, 12, 71–78. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.R.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Distefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Metab. Pathw. Eng. 2018, 1706, 91–110. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Song, S.; Wu, Z.; Wang, C.; Liu, B.; Ye, X.; Chen, J.; Yang, Q.; Ye, H.; Xu, B.; Wang, L. RCCRT1 Is Correlated With Prognosis and Promotes Cell Migration and Invasion in Renal Cell Carcinoma. Urology 2014, 84, 730.e1. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Bai, H.; Dang, Y.; Lv, P.; Wu, S. Long intergenic non-coding RNA 00152 promotes renal cell carcinoma progression by epigenetically suppressing P16 and negatively regulates miR-205. Am. J. Cancer Res. 2017, 7, 312–322. [Google Scholar] [PubMed]
- Xue, D.; Wang, H.; Chen, Y.; Shen, D.; Lu, J.; Wang, M.; Zebibula, A.; Xu, L.; Wu, H.; Li, G.; et al. Circ–AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR–296–3p/E–cadherin signals. Mol. Cancer 2019, 18, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattick, J.W.; Rinn, J. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Boil. 2015, 22, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Thrash-Bingham, C.A.; Tartof, K.D. aHIF: A Natural Antisense Transcript Overexpressed in Human Renal Cancer and During Hypoxia. J. Natl. Cancer Inst. 1999, 91, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi, D.; Iurlaro, R.; Sordet, O.; Marinello, J.; Zaffaroni, N.; Capranico, G. Characterization of novel antisense HIF-1α transcripts in human cancers. Cell Cycle 2011, 10, 3189–3197. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, J.; Alam, J.; Rothenburg, J.; Deng, M.; Schmidt, D.; Syring, I.; Miersch, H.; Perner, S.; Müller, S.C. The long non–coding RNA lnc–ZNF180–2 is a prognostic biomarker in patients with clear cell renal cell carcinoma. Am. J. Cancer Res. 2015, 5, 2799–2807. [Google Scholar]
- Ren, X.; Lan, T.; Chen, Y.; Shao, Z.; Yang, C.; Peng, J. lncRNA uc009yby.1 promotes renal cell proliferation and is associated with poor survival in patients with clear cell renal cell carcinomas. Oncol. Lett. 2016, 12, 1929–1934. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Han, Z.; Qian, J.; Bao, M.; Li, P.; Ju, X.; Zhang, S.; Zhang, L.; Li, S.; Cao, Q.; et al. Expression Pattern of Long Non-Coding RNAs in Renal Cell Carcinoma Revealed by Microarray. PLoS ONE 2014, 9, e99372. [Google Scholar] [CrossRef] [Green Version]
- Blondeau, J.J.; Deng, M.; Syring, I.; Schrödter, S.; Schmidt, D.; Perner, S.; Müller, S.C.; Ellinger, J. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clin. Epigenetics 2015, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhang, C.; Xia, S.-Y.; Xiu, Y.-C.; Yan, H.-Y. Downregulation of long non-coding RNA TRIM52-AS1 functions as a tumor suppressor in renal cell carcinoma. Mol. Med. Rep. 2016, 13, 3206–3212. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, Y.; Chen, D.; Yu, Z.; Ni, L.; Mao, X.; Gui, Y.; Lai, Y.; Wang, T.; Jin, L.; et al. Identification of long-non coding RNA UCA1 as an oncogene in renal cell carcinoma. Mol. Med. Rep. 2016, 13, 3326–3334. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.-Q.; Weng, W.-W.; Zhang, Q.-Y.; Yang, X.-Q.; Gan, H.-L.; Yang, Y.-S.; Zhang, P.-P.; Sun, M.-H.; Xu, M.-D.; et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis 2016, 5, e192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.-H.; Qin, X.-H.; Zhang, X.-L.; Yi, J.-W.; Han, J.-Y. Long noncoding RNA GIHCG is a potential diagnostic and prognostic biomarker and therapeutic target for renal cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 46–54. [Google Scholar] [PubMed]
- Qu, L.; Wu, Z.; Li, Y.; Xu, Z.; Liu, B.; Liu, F.; Bao, Y.; Wu, D.; Liu, J.; Wang, A.; et al. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat. Commun. 2016, 7, 12692. [Google Scholar] [CrossRef]
- Bao, X.; Duan, J.; Yan, Y.; Ma, X.; Zhang, Y.; Wang, H.; Ni, D.; Wu, S.; Peng, C.; Fan, Y.; et al. Upregulation of long noncoding RNA PVT1 predicts unfavorable prognosis in patients with clear cell renal cell carcinoma. Cancer Biomark. 2017, 21, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Bao, Y.; Wu, Z.; Zhao, T.; Wang, D.; Shi, J.; Liu, B.; Sun, S.; Yang, F.; Wang, L.; et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Chen, Y.; Wang, Y.; Liu, S.; Yuan, X.; Pan, F.; Geng, P. Decreased expression of a novel lncRNA CADM1-AS1 is associated with poor prognosis in patients with clear cell renal cell carcinomas. Int. J. Clin. Exp. Pathol. 2014, 7, 2758–2767. [Google Scholar]
- Zhang, H.-M.; Yang, F.-Q.; Yan, Y.; Che, J.-P.; Zheng, J.-H. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 5801–5809. [Google Scholar]
- Xue, S.; Li, Q.-W.; Che, J.-P.; Guo, Y.; Yang, F.-Q.; Zheng, J.-H. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 3765–3774. [Google Scholar]
- Zhang, H.-M.; Yang, F.-Q.; Chen, S.-J.; Che, J.; Zheng, J.-H. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumor Boil. 2014, 36, 2947–2955. [Google Scholar] [CrossRef]
- Wang, L.; Cai, Y.; Zhao, X.; Jia, X.; Zhang, J.; Liu, J.; Zhen, H.; Wang, T.; Tang, X.; Liu, Y.; et al. Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma 2015, 62, 412–418. [Google Scholar] [CrossRef]
- Wu, Y.; Tan, C.; Weng, W.-W.; Deng, Y.; Zhang, Q.-Y.; Yang, X.-Q.; Gan, H.-L.; Wang, T.; Zhang, P.-P.; Xu, M.-D.; et al. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am. J. Cancer Res. 2016, 6, 285–299. [Google Scholar] [PubMed]
- Wang, P.-Q.; Wu, Y.-X.; Zhong, X.-D.; Liu, B.; Qiao, G. Prognostic significance of overexpressed long non-coding RNA TUG1 in patients with clear cell renal cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 82–86. [Google Scholar] [PubMed]
- Su, H.; Sun, T.; Wang, H.; Shi, G.; Zhang, H.; Sun, F.-K.; Ye, D. Decreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinoma. Oncotarget 2016, 8, 5789–5799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, X.; Siprashvili, Z.; Eminaga, O.; Shen, Z.; Sato, Y.; Kume, H.; Homma, Y.; Ogawa, S.; Khavari, P.A.; Pollack, J.R.; et al. Novel lincRNA SLINKY is a prognostic biomarker in kidney cancer. Oncotarget 2017, 8, 18657–18669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Tong, Y.; Zhu, J.; Lei, Z.; Wan, L.; Zhu, X.; Ye, F.; Xie, L. An increase in long non-coding RNA PANDAR is associated with poor prognosis in clear cell renal cell carcinoma. BMC Cancer 2017, 17, 373. [Google Scholar] [CrossRef]
- Li, J.-K.; Chen, C.; Liu, J.-Y.; Shi, J.-Z.; Liu, S.-P.; Liu, B.; Wu, D.-S.; Fang, Z.-Y.; Bao, Y.; Jiang, M.-M.; et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol. Cancer 2017, 16, 111. [Google Scholar] [CrossRef]
- Yang, T.; Zhou, H.; Liu, P.; Yan, L.; Yao, W.; Chen, K.; Zeng, J.; Li, H.; Hu, J.; Xu, H.; et al. lncRNA PVT1 and its splicing variant function as competing endogenous RNA to regulate clear cell renal cell carcinoma progression. Oncotarget 2017, 8, 85353–85367. [Google Scholar] [CrossRef] [Green Version]
- Flippot, R.; Mouawad, R.; Spano, J.-P.; Rouprêt, M.; Compérat, E.; Bitker, M.-O.; Parra, J.; Vaessen, C.; Allanic, F.; Manach, Q.; et al. Expression of long non-coding RNA MFI2-AS1 is a strong predictor of recurrence in sporadic localized clear-cell renal cell carcinoma. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Su, H.; Wang, H.; Shi, G.-H.; Zhang, H.; Sun, F.; Ye, D. Downregulation of long non-coding RNA ENSG00000241684 is associated with poor prognosis in advanced clear cell renal cell carcinoma. Eur. J. Surg. Oncol. 2018, 44, 840–846. [Google Scholar] [CrossRef]
- Wang, L.-N.; Zhu, X.-Q.; Song, X.-S.; Xu, Y. Long noncoding RNA lung cancer associated transcript 1 promotes proliferation and invasion of clear cell renal cell carcinoma cells by negatively regulating miR–495–3p. J. Cell. Biochem. 2018, 119, 7599–7609. [Google Scholar] [CrossRef]
- Ding, C.; Han, F.; Xiang, H.; Xia, X.; Wang, Y.; Dou, M.; Zheng, J.; Li, Y.; Xue, W.; Ding, X.; et al. LncRNA CRNDE is a biomarker for clinical progression and poor prognosis in clear cell renal cell carcinoma. J. Cell. Biochem. 2018, 119, 10406–10414. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Ding, J.; Chen, C.; Wu, Z.; Liu, B.; Gao, Y.; Chen, W.; Liu, F.; Sun, W.; Li, X.-F.; et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016, 29, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yang, F.; Wei, D.; Liu, B.; Chen, C.; Bao, Y.; Wu, Z.; Wu, D.; Tan, H.; Li, J.; et al. Long noncoding RNA–SRLR elicits intrinsic sorafenib resistance via evoking IL–6/STAT3 axis in renal cell carcinoma. Oncogene 2017, 36, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Sun, Y.; Guo, C.; Hu, G.; Wang, M.; Zheng, J.; Lin, W.; Huang, Q.; Li, G.; Zheng, J.; et al. LncRNA–SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA–143–3p signals. Cell Death Differ. 2017, 24, 1502–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Pang, X.; Shang, W.; Xie, H.; Feng, Y.; Feng, G. Long non–coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR–21/SOX5 pathway. Cell Cycle 2019, 18, 257–263. [Google Scholar] [CrossRef] [PubMed]


| Year | Diagnostic Biomarker | Biological Source | Number of Cases/Controls | Diagnostic Performance | Reference | ||
|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | AUC | |||||
| 2009 | miR-200c | Tissue | 72 ccRCC; 72 MNT | n.a. | n.a. | 0.970 | [56] |
| 2010 | miR-200c | Tissue | 13 chRCC; 21 oncocytomas | n.a. | n.a. | 0.880 | [57] |
| 2012 | miR-21 | Tissue | 71 ccRCC & 18 pRCC; 10 chRCC & 8 oncocytomas | 83.0 | 90.0 | 0.886 | [58] |
| 2013 | 3 miR panel | Tissue | 111 ccRCC; 111 MNT | 95.0 | 100.0 | 0.990 | [54] |
| 2013 | miR-210 + let-7c | Tissue | 16 pRCC type I; 17 pRCC type II | n.a. | n.a. | 0.919 | [59] |
| 2013 | miR-200b | Tissue | 90 RCC; 30 oncocytomas | 96.7 | 90.0 | 0.914 | [55] |
| 2014 | miR-3687 | Tissue | 24 ccRCC; 40 NRT | n.a. | n.a. | 0.847 | [60] |
| 2014 | miR-141 | Tissue | 68 ccRCC; 68 MNT | 86.8 | 97.1 | 0.930 | [61] |
| 2014 | miR-129-3p | Tissue | 69 ccRCC; 69 MNT | 75.9 | 62.1 | 0.735 | [62] |
| 2014 | 5 miR panel | Tissue | 32 ccRCC; 16 NRT | 100.0 | 100.0 | 1.000 | [63] |
| 2015 | 3 piRNA panel | Tissue | 106 ccRCC; 77 NRT | 91.0 | 86.0 | 0.910 | [51] |
| 2016 | miR-145 | Tissue | 44 RCC; 44 MNT | n.a. | n.a. | 0.616 | [64] |
| 2016 | miR-141 | Tissue | 27 ccRCC; 27 MNT | n.a. | n.a. | 0.912 | [65] |
| 2016 | piR-823 | Tissue | 153 RCC; 121 MNT | n.a. | n.a. | 0.795 | [53] |
| 2017 | 4 miR panel | Tissue | 48 ccRCC; 50 benign renal tumors | 91.7 | 94.0 | 0.992 | [66] |
| 2017 | miR-34a | Tissue | 85 RCC; 85 MNT | n.a. | n.a. | 0.854 | [67] |
| 2017 | miR-200c | Tissue | 19 chRCC; 11 oncocytomas | 84.0 | 82.0 | 0.820 | [68] |
| 2017 | miR-200c | Tissue | 30 ccRCC; 30 MNT | n.a. | n.a. | 0.860 | [69] |
| 2017 | miR-720 | Tissue | 30 RCC; 30 NRT | 80.0 | 100.0 | 0.905 | [70] |
| 2018 | miR-203 | Tissue | 53 ccRCC; 53 MNT | n.a. | n.a. | 0.944 | [71] |
| 2018 | miR-182-5p | Tissue | 24 ccRCC; 24 MNT | n.a. | n.a. | 0.954 | [72] |
| 2018 | miR-224/miR-141 | Tissue | 68 ccRCC; 68 MNT | 97.1 | 98.5 | 0.990 | [73] |
| 2018 | miR-452-5p | Tissue | 20 RCC; 20 MNT | n.a. | n.a. | 0.919 | [74] |
| 2019 | piR-34536 | Tissue | 118 ccRCC; 75 NRT | 78.0 | 78.1 | 0.815 | [75] |
| Year | Diagnostic Biomarker | Biological Source | Number of Cases/Controls | Diagnostic Performance | Reference | ||
|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | AUC | |||||
| 2011 | miR-1233 | Serum | 84 RCC; 93 AC | 77.4 | 37.6 | 0.588 | [77] |
| 2012 | miR-378 + miR-451 | Serum | 90 RCC; 35 AC | 81.0 | 83.0 | 0.860 | [84] |
| 2013 | miR-210 | Serum | 68 ccRCC; 42 AC | 81.0 | 79.4 | 0.874 | [78] |
| 2014 | miR-210 | Serum | 34 ccRCC; 23 AC | 65.0 | 83.0 | 0.770 | [85] |
| 2014 | miR-221 | Plasma | 43 RCC; 34 AC | 72.5 | 33.3 | 0.696 | [86] |
| 2015 | 5 miR panel | Serum | 76 stage I ccRCC; 107 AC | 80.0 | 71.0 | 0.807 | [87] |
| 2015 | miR-210 + miR-378 | Serum | 195 RCC; 100 AC | 80.0 | 78.0 | 0.848 | [88] |
| 2016 | miR-126-3p + miR-486-5p | Urine exosomes | 24 benign renal tumors; 33 AC | 75.0 | 87.5 | 0.850 | [82] |
| 2016 | piR-823 | Serum | 178 RCC; 101 AC | n.a. | n.a. | 0.626 | [53] |
| Urine | 20 RCC; 15 AC | n.a. | n.a. | 0.743 | |||
| 2017 | miR-144-3p | Plasma | 106 ccRCC; 123 AC | 87.1 | 83.0 | 0.910 | [81] |
| 2017 | miR-21 | Plasma | 30 ccRCC; 30 AC | 77.3 | 96.4 | 0.865 | [80] |
| 2017 | miR-210 | Urine | 75 ccRCC; 45 AC | 57.8 | 80.0 | 0.760 | [89] |
| 2017 | Let-7a | Urine | 69 ccRCC; 36 AC | 71.0 | 81.0 | 0.831 | [90] |
| 2017 | miR-451 | Plasma | 94 ccRCC; 100 AC | n.a. | n.a. | 0.640 | [91] |
| 2018 | miR-1233 | Serum exosomes | 80 ccRCC; 82 AC | 81.0 | 76.0 | 0.820 | [79] |
| 2018 | miR-15a | Urine | 67 RCT; 15 AC | 98.1 | 100 | 0.955 | [83] |
| 2018 | miR-210 × miR-224 | Plasma | 66 ccRCC; 67 AC | 92.5 | 45.5 | 0.659 | [73] |
| 2018 | miR-210 | Serum exosomes | 45 ccRCC; 30 AC | 82.5 | 80.0 | 0.878 | [92] |
| 2019 | miR-508-3p & miR-885-5p | Serum | 85 ccRCC; 35 AC | n.a. | n.a. | 0.900 | [93] |
| 2020 | miR-432-5p | Urine | 44 ccRCC-SRM; 27 oncocytomas | n.a. | n.a. | 0.710 | [94] |
| 2020 | miR-30a-5pme | Urine | 171 ccRCC; 85 AC | 63.0 | 67.0 | 0.670 | [95] |
| Year | Prognostic Variable | Prognostic Biomarker | Biological Source | Number of Cases | Poor Prognosis | Prognostic Performance | Reference | |
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | |||||||
| 2010 | RFS | miR-9-3 | Tissue | 59 ccRCC | High methylation | 5.850 | 1.300–26.35 | [104] |
| 2012 | CSS | 4 miR panel | Tissue | 68 ccRCC | High risk | 8.800 * | 2.620–29.58 * | [105] |
| 2012 | DFS | miR-21 | Tissue | 87 RCC | Positive expression | 2.150 * | 1.160–3.980 * | [58] |
| 2013 | RFS | miR-124-3 | Tissue | 80 ccRCC | High methylation | 9.370 | 2.680–32.80 | [106] |
| 2013 | RFS | miR-514 | Tissue | 87 ccRCC | Low expression | 0.250 | 0.080–0.750 | [54] |
| 2013 | OS | miR-210 | Tissue | 46 ccRCC | Low expression | 3.010 | 1.390–6.510 | [101] |
| 2013 | CSS | miR-486 | Tissue | 46 RCC | High expression | 4.330 | 1.450–18.71 | [107] |
| 2013 | OS | miR–100 | Tissue | 96 RCC | High expression | 3.600 | 1.800–5.200 | [96] |
| 2013 | CSS | miR–155 | Tissue | 137 ccRCC | Low expression | 5.490 | 2.400–12.52 | [103] |
| 2014 | DFS | miR–21 & miR–126 | Tissue | 103 ccRCC | High risk | 19.37 | 4.060–92.44 | [108] |
| 2014 | OS | miR–630 | Tissue | 92 ccRCC | High expression | 3.021 | 2.074–5.726 | [97] |
| 2014 | DSS | miR–21/miR–10b | Tissue | 105 ccRCC | High ratio | 2.624 | 1.201–5.736 | [109] |
| 2014 | DFS | miR–129–3p | Tissue | 48 ccRCC | Low expression | 3.119 | 1.060–9.175 | [62] |
| 2014 | RFS | miR–125b | Tissue | 200 ccRCC | High expression | 3.931 | 1.213–12.74 | [110] |
| 2015 | OS | miR–497 | Tissue | 86 ccRCC | Low expression | 2.583 | 1.691–6.361 | [111] |
| 2015 | DFS | miR–210 | Tissue | 258 ccRCC | Positive expression | 1.910 | 1.010–3.310 | [98] |
| 2015 | DFS | miR–126 | Tissue | 260 ccRCC | Negative expression | 0.300 * | 0.180–0.500 * | [112] |
| 2015 | OS | miR–506 | Tissue | 106 ccRCC | Low expression | 3.886 | 2.179–7.524 | [113] |
| 2015 | OS | miR–203 | Tissue | 90 ccRCC | Low expression | 3.071 | 1.719–6.374 | [114] |
| 2015 | CSS | miR–21 | Tissue | 45 RCC | High expression | 6.460 | 1.350–30.94 | [115] |
| 2015 | CSS | piR–43607 | Tissue | 68 ccRCC | High expression | 1.240 * | 1.082–1.445 * | [52] |
| 2015 | OS | miR–124–3p | Tissue | 62 ccRCC | Low expression | 2.600 * | 1.069–7.262 * | [116] |
| 2015 | RFS | piR–38756 | Tissue | 72 ccRCC | High expression | 3.150 | 1.960–9.320 | [51] |
| 2015 | PFS | miR–27a–3p | Tissue | 140 ccRCC | High expression | 2.710 | 1.230–6.420 | [102] |
| 2016 | DFS | miR–194 | Tissue | 234 ccRCC | Negative expression | 0.520 | 0.270–0.980 | [117] |
| 2016 | DFS | miR–19a | Tissue | 197 ccRCC | High expression | 2.410 | 1.217–4.773 | [118] |
| 2016 | PFS | miR–222–3p | Tissue | 74 ccRCC | High expression | 2.020 | 1.510–2.710 | [119] |
| 2017 | CSS | miR–223–3p | Tissue | 78 ccRCC | High expression | 3.510 | 1.600–7.690 | [66] |
| 2017 | DFS | miR–10b | Tissue | 246 ccRCC | Negative expression | 0.470 * | 0.280–0.790 * | [120] |
| 2017 | OS | miR–766–3p | Tissue | 75 RCC | Low expression | 2.700 | 1.310–5.530 | [121] |
| 2018 | OS | miR–566 | Tissue | 42 ccRCC | High expression | 0.060 | 0.005–0.769 | [122] |
| 2018 | OS | miR–18–5p | Tissue | 42 RCC | High expression | 0.175 | 0.032–0.953 | [123] |
| 2018 | OS | miR–663a | Tissue | 42 ccRCC | High expression | 5.132 | 1.039–25.350 | [124] |
| 2018 | OS | miR–572 | Tissue | 42 RCC | High expression | 0.174 | 0.034–0.878 | [125] |
| 2018 | RFS | miR–155–5p & miR–210–3p | Tissue | 205 ccRCC | High risk | 2.700 | 1.280–5.680 | [100] |
| 2018 | OS | miR–452–5p | Tissue | 102 RCC | High expression | 1.580 | 1.070–2.310 | [74] |
| 2019 | OS | miR–23a–5p | Tissue | 118 RCC | High expression | 3.270 | 1.552–6.893 | [126] |
| 2019 | OS | miR–183–5p | Tissue | 284 ccRCC | High expression | 0.550 | 0.364–0.832 | [127] |
| 2019 | OS | miR–3133 | Tissue | 135 ccRCC | Low expression | 2.802 | 1.391–5.646 | [128] |
| 2019 | OS | miR–221–5p | Tissue | 196 ccRCC | High expression | 0.550 | 0.326–0.926 | [129] |
| 2019 | PFS | piR–51810 | Tissue | 118 ccRCC | Low expression | 0.431 | 0.190–0.975 | [75] |
| 2019 | OS | miR–142–3p | Tissue | 284 RCC | High expression | 0.525 | 0.347–0.796 | [130] |
| 2019 | OS | miR–106b–5p | Tissue | 284 ccRCC | High expression | 0.496 | 0.327–0.752 | [131] |
| 2019 | MFS | 4 miR panel | Tissue | 85 ccRCC | High risk | 12.402 | 3.586–42.893 | [132] |
| 2020 | DFS | miR–30a–5pme | Tissue | 227 ccRCC | High methylation | 5.174 | 1.228–21.808 | [95] |
| Year | Prognostic Variable | Prognostic Biomarker | Biological Source | Number of Cases | Poor Prognosis | Prognostic Performance | Reference | |
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | |||||||
| 2014 | CSS | miR–221 | Plasma | 43 RCC | High expression | 10.70 | 1.330–85.65 | [86] |
| 2017 | OS | let–7i–5p | Plasma exosomal | 65 RCC | Low expression | 0.566 | 0.374–0.857 | [133] |
| 2017 | CSS | miR–210 & miR–221 & miR–1233 | Plasma | 50 ccRCC | High risk | 3.890 | 1.260–12.01 | [99] |
| 2017 | PFS | miR–224 | Plasma exosomal | 108 ccRCC | High expression | 11.00 | 3.300–68.70 | [134] |
| 2017 | DSS | miR–150 | Plasma | 94 ccRCC | Low expression | 1.280 | 1.020–1.670 | [91] |
| 2018 | PFS | miR–206 | Serum | 67 ccRCC | High expression | 3.670 | 1.290–10.51 | [135] |
| 2020 | OS | miR–328–3p | Urine | 44 ccRCC | Low expression | 0.290 | 0.080–1.030 | [94] |
| 2020 | DSS | miR–30a–5pme | Urine | 53 ccRCC | High methylation | 9.376 | 1.158–75.903 | [95] |
| Year | Predictive Biomarker | Biological Source | Number of Cases/Cell Lines | Type of Therapy | Main Findings | Reference |
|---|---|---|---|---|---|---|
| 2012 | multi–miR panels | Whole blood | 38 ccRCC | Targeted therapy | Several miRs ⇒ prolonged or poor response to sunitinib | [146] |
| 2013 | miR–141 | Tissue/in vitro | 20 ccRCC | Targeted Therapy | ↓ miR–141 ⇒ poor response to sunitinib | [143] |
| 2013 | miR–381 | In vitro | 786–O | Chemotherapy | MiR–381 + 5–FU ⇒ lower proliferation, and ↑ 5–FU efficacy | [147] |
| 2014 | miR–942 | Tissue/in vitro | 20 RCC & Caki–2 | Targeted Therapy | MiR–942 ⇒ sunitinib resistance ⇒ ↓ TTP & OS | [148] |
| 2014 | miR–200c | In vitro | HEK293, SN12C, ACHN, 786–O & Caki–1 | Targeted Therapy | Mimic miR–200c ⇒ sensitivity to therapy with TKI | [149] |
| 2015 | miR–27b | In vitro/in vivo | ACHN, 769–P, 786–O & Caki–1 | Chemotherapy | Overexpressing miR–27b ⇒ sensitizes RCC cells to a variety of anti–cancer drugs, such as doxorubicin | [150] |
| 2015 | miR–30a | Tissue/in vivo | 10 ccRCC & A498 + 786–O | Targeted Therapy | Exogenously expression of miR–30a ⇒ ↑ sorafenib treatment efficacy | [151] |
| 2015 | miR–200c | In vitro | Caki–1, Caki–2, A498, ACHN, 786–O & 769–P | Chemotherapy | ↓ miR–200c ⇒ resistance to docetaxel | [152] |
| 2015 | miR–155 & miR–484 | Tissue | 63 RCC | Targeted Therapy | ↓ of both miRs ⇒ better response to sunitinib ⇒ ↑ TTP | [144] |
| 2015 | miR–124 | In vitro | Caki–2 | Chemotherapy | ↓ miR–124 ⇒ resistance to doxorubicin and vinblastine | [153] |
| 2015 | miR–221 & miR–222 | Tissue/in vivo | 30 ccRCC & 786–O + ACHN | Targeted Therapy | ↑ of both miRs ⇒ poor response to sunitinib therapy | [154] |
| 2016 | miR–99b–5p | Tissue | 40 ccRCC | Targeted Therapy | ↓ miR–99b–5p ⇒ ↓ PFS and in TKI non–responders | [155] |
| 2017 | miR–605 | Serum | 36 ccRCC | Targeted Therapy | MiR–605 ⇒ ↓ after vorinostato and bevacizumab therapy in responders | [156] |
| 2017 | miR–27b & miR–23b & miR–628–5p | Tissue | 123 RCC | Targeted Therapy | ↑ of these miRs ⇒ long–term sunitinib response | [157] |
| 2017 | miR–144–3p | In vitro/in vivo | 786–O & SN12–PM6 + Nude mice | Targeted Therapy | ↑ miR–144–3p ⇒ ↓ ARID1A and resistance to sunitinib | [158] |
| 2017 | miR–451 | In vitro | ACHN & GRC–1 | Chemotherapy | MiR–451 knockdown ⇒ ↑ sensitivity to adriamycin therapy | [159] |
| 2018 | miR–942 & miR–133 | Tissue | 56 RCC | Targeted Therapy | Both miRs ⇒ discriminate between sunitinib responders and non–responders | [160] |
| 2019 | miR–421 | Tissue | 101 MRCC | Targeted Therapy | ↑ miR–421 in TKI non–responders | [145] |
| 2019 | miR–376b–3p | Tissue | 132 ccRCC | Targeted Therapy | ↓ miR–376b–3p in sunitinib poor responders | [161] |
| 2020 | miR–31–5p | Exosomes from plasma/in vitro/in vivo | 40 PD MRCC + 786–O + BALB/c nude mice | Targeted Therapy | ↑ miR–31–5p in PD vs non–PD patients’ plasma samples treated with sorafenib | [162] |
| Year | Diagnostic Biomarker | Biological Source | Number of Cases/Controls | Diagnostic Performance | Reference | ||
|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | AUC | |||||
| 1999 | aHIF | Tissue | 10 ccRCC; 7 pRCC | n.a. | n.a. | n.a. | [175] |
| 2011 | aHIF | Tissue | 26 RCC; 26 MNT | n.a. | n.a. | n.a. | [176] |
| 2014 | AK096725 | Tissue | 70 RCC; 70 MNT | n.a. | n.a. | n.a. | [179] |
| 2015 | TTC34–3 | Tissue | 55 ccRCC; 52 NRT | n.a. | n.a. | 0.990 | [180] |
| 2015 | CYP4A22–2/3 | Tissue | 102 ccRCC; 50 NRT | n.a. | n.a. | 0.790 | [177] |
| 2016 | TRIM52–AS1 | Tissue | 60 RCC; 60 MNT | n.a. | n.a. | n.a. | [181] |
| 2016 | UCA1 | Tissue | 46 RCC; 46 MNT | n.a. | n.a. | n.a. | [182] |
| 2016 | UC009YBY.1 | Tissue | 70 RCC; 70 MNT | 54.3 | 82.9 | 0.700 | [178] |
| 2018 | HOTAIR | Tissue | 24 ccRCC; 24 MNT | n.a. | n.a. | 0.923 | [70] |
| 2016 | 5 lncRNA panel | Serum | 24 ccRCC; 27 AC | 79.2 | 88.9 | 0.900 | [183] |
| 2018 | GIHCG | Serum | 31 Stage I ccRCC; 46 AC | 80.7 | 84.8 | 0.886 | [184] |
| Year | Prognostic Variable | Prognostic Biomarker | Biological Source | Number of Cases | Poor Prognosis | Prognostic Performance | Reference | |
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | |||||||
| 2014 | OS | CADM1–AS1 | Tissue | 64 ccRCC | Low expression | 0.211 | 0.088–0.504 | [188] |
| 2014 | OS | SPRY4–IT1 | Tissue | 98 ccRCC | High expression | 3.375 | 1.824–7.391 | [189] |
| 2015 | OS | NBAT–1 | Tissue | 98 ccRCC | Low expression | 3.701 | 1.261–9.784 | [190] |
| 2015 | OS | MALAT1 | Tissue | 106 ccRCC | High expression | 3.086 | 1.813–7.025 | [191] |
| 2015 | OS | H19 | Tissue | 92 ccRCC | High expression | 3.894 | 1.872–8.014 | [192] |
| 2015 | PFS | ZNF180–2 | Tissue | 102 ccRCC | Low expression | 0.803 | 0.699–0.922 | [177] |
| 2016 | OS | Linc00152 | Tissue | 77 ccRCC | High expression | 2.577 | 1.233–5.387 | [193] |
| 2016 | RFS | lncARSR | Tissue | 205 ccRCC | High expression | 2.023 | 1.213–3.375 | [185] |
| 2017 | OS | TUG1 | Tissue | 203 ccRCC | High expression | 2.337 | 1.451–6.673 | [194] |
| 2017 | OS | TCL6 | Tissue | 71 ccRCC | Low expression | 0.130 | 0.020–0.680 | [195] |
| 2017 | OS | SLINKY | Tissue | 100 ccRCC | High expression | 8.440 | 1.770–40.23 | [196] |
| 2017 | OS | PANDAR | Tissue | 62 ccRCC | High expression | 1.130 | 0.980–5.120 | [197] |
| 2017 | OS | MRCCAT1 | Tissue | 68 ccRCC | High expression | 2.306 | 1.003–2.849 | [198] |
| 2017 | OS | PVT1 | Tissue | 50 ccRCC | High expression | 1.494 | 1.081–2.063 | [199] |
| 2017 | DFS | MFI2–AS1 | Tissue | 167 ccRCC | Positive expression | 4.240 | 2.070–8.700 | [200] |
| 2017 | DFS | PVT1 | Tissue | 129 ccRCC | High expression | 3.553 | 1.515–8.329 | [186] |
| 2018 | OS | ENSG00000241684 | Tissue | 61 ccRCC | Low expression | 5.378 | 2.084–13.88 | [201] |
| 2018 | OS | LUCAT1 | Tissue | 64 ccRCC | High expression | 3.650 | 1.356–9.826 | [202] |
| 2018 | OS | CRNDE | Tissue | 112 ccRCC | High expression | 2.023 | 1.039–3.468 | [203] |
| 2019 | OS | EGFR–AS1 | Tissue | 204 RCC | High expression | 2.204 | 1.145–4.241 | [187] |
| 2016 | PFS | lncARSR | Plasma | 71 ccRCC | High expression | n.a. | n.a. | [204] |
| 2018 | OS | GIHCG | Serum | 46 ccRCC | High expression | n.a. | n.a. | [184] |
| Year | Predictive Biomarker | Biological Source | Number of Cases/Cell Lines | Type of Therapy | Main Findings | Reference |
|---|---|---|---|---|---|---|
| 2016 | lncARSR | Plasma | 71 ccRCC | TargetedTherapy | ↑ lncARSR in progressive disease during sunitinib therapy | [204] |
| 2017 | lncSRLR | Tissue/in vitro | 51 RCC; RCC cell lines | TargetedTherapy | ↑ lncSRLR in sorafenib non–responders. In vitro ablation ⇒ ↑ therapeutic sensitivity | [205] |
| 2017 | SARCC | In vitro | RCC cell lines | TargetedTherapy | ↑ SARCC after sunitinib treatment ⇒ ↓ sunitinib resistance (positive feedback loop) | [206] |
| 2019 | GAS5 | Tissue/ in vitro/ in vivo | 15 ccRCC; RCC cell lines; mice model | TargetedTherapy | ↓ GAS5 in sorafenib non–responders; in vitro/ in vivo re–introduction ⇒ ↑ sensitivity | [207] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Outeiro-Pinho, G.; Barros-Silva, D.; Correia, M.P.; Henrique, R.; Jerónimo, C. Renal Cell Tumors: Uncovering the Biomarker Potential of ncRNAs. Cancers 2020, 12, 2214. https://doi.org/10.3390/cancers12082214
Outeiro-Pinho G, Barros-Silva D, Correia MP, Henrique R, Jerónimo C. Renal Cell Tumors: Uncovering the Biomarker Potential of ncRNAs. Cancers. 2020; 12(8):2214. https://doi.org/10.3390/cancers12082214
Chicago/Turabian StyleOuteiro-Pinho, Gonçalo, Daniela Barros-Silva, Margareta P. Correia, Rui Henrique, and Carmen Jerónimo. 2020. "Renal Cell Tumors: Uncovering the Biomarker Potential of ncRNAs" Cancers 12, no. 8: 2214. https://doi.org/10.3390/cancers12082214
APA StyleOuteiro-Pinho, G., Barros-Silva, D., Correia, M. P., Henrique, R., & Jerónimo, C. (2020). Renal Cell Tumors: Uncovering the Biomarker Potential of ncRNAs. Cancers, 12(8), 2214. https://doi.org/10.3390/cancers12082214