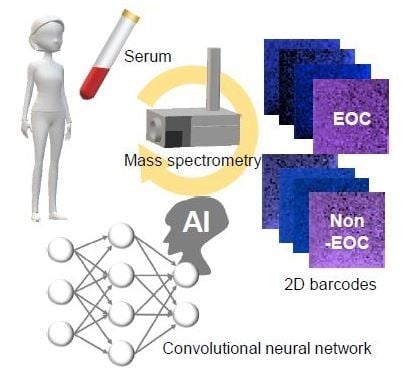
Comprehensive Serum Glycopeptide Spectra Analysis Combined with Artificial Intelligence (CSGSA-AI) to Diagnose Early-Stage Ovarian Cancer
Abstract
:1. Introduction
2. Results
2.1. The Training Options
2.2. Learning Efficacy of Pretrained CNN Models
2.3. Glycopeptide Alignment to Generate 2D Barcodes
2.4. Training with 2D Barcodes Colored by CA125 and HE4
2.5. CSGSA-AI Scheme and its Diagnostic Performance
3. Discussion
4. Materials and Methods
4.1. Patient Samples
4.2. Study Approval
4.3. Sample Preparation
4.4. Liquid Chromatography and Mass Spectrometry
4.5. Generation of 2D Barcodes
4.6. Training CNN to Distinguish Between EOC and Non-EOC Patterns
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ward, E.M.; Sherman, R.L.; Henley, S.J.; Jemal, A.; Siegel, D.A.; Feuer, E.J.; Firth, A.U.; Kohler, B.A.; Scott, S.; Ma, J.; et al. Annual Report to the Nation on the Status of Cancer, Featuring Cancer in Men and Women Age 20–49 Years. J. Natl. Cancer Inst. 2019, 111, 1279–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Henley, S.J.; Ward, E.M.; Scott, S.; Ma, J.; Anderson, R.N.; Firth, A.U.; Thomas, C.C.; Islami, F.; Weir, H.K.; Lewis, D.R.; et al. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer 2020, 126, 2225–2249. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Ward, E.M.; Johnson, C.J.; Cronin, K.A.; Ma, J.; Ryerson, B.; Mariotto, A.; Lake, A.J.; Wilson, R.; Sherman, R.L.; et al. Annual Report to the Nation on the Status of Cancer, 1975–2014, Featuring Survival. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Cancer Stat Facts: Ovarian Cancer. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 10 May 2020).
- Khiewvan, B.; Torigian, D.A.; Emamzadehfard, S.; Paydary, K.; Salavati, A.; Houshmand, S.; Werner, T.J.; Alavi, A. An update on the role of PET/CT and PET/MRI in ovarian cancer. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1079–1091. [Google Scholar] [CrossRef]
- Sironi, S.; Messa, C.; Mangili, G.; Zangheri, B.; Aletti, G.; Garavaglia, E.; Vigano, R.; Picchio, M.; Taccagni, G.; Maschio, A.D.; et al. Integrated FDG PET/CT in patients with persistent ovarian cancer: Correlation with histologic findings. Radiology 2004, 233, 433–440. [Google Scholar] [CrossRef]
- Meany, D.L.; Sokoll, L.J.; Chan, D.W. Early Detection of Cancer: Immunoassays for Plasma Tumor Markers. Expert Opin. Med. Diagn. 2009, 3, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Dorigo, O.; Berek, J.S. Personalizing CA125 levels for ovarian cancer screening. Cancer Prev. Res. 2011, 4, 1356–1359. [Google Scholar] [CrossRef] [Green Version]
- Buys, S.S.; Partridge, E.; Black, A.; Johnson, C.C.; Lamerato, L.; Isaacs, C.; Reding, D.J.; Greenlee, R.T.; Yokochi, L.A.; Kessel, B.; et al. Effect of screening on ovarian cancer mortality: The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Randomized Controlled Trial. JAMA 2011, 305, 2295–2303. [Google Scholar] [CrossRef]
- Pinsky, P.F.; Yu, K.; Kramer, B.S.; Black, A.; Buys, S.S.; Partridge, E.; Gohagan, J.; Berg, C.D.; Prorok, P.C. Extended mortality results for ovarian cancer screening in the PLCO trial with median 15 years follow-up. Gynecol. Oncol. 2016, 143, 270–275. [Google Scholar] [CrossRef] [Green Version]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.I.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef] [PubMed]
- Mari, R.; Mamessier, E.; Lambaudie, E.; Provansal, M.; Birnbaum, D.; Bertucci, F.; Sabatier, R. Liquid Biopsies for Ovarian Carcinoma: How Blood Tests May Improve the Clinical Management of a Deadly Disease. Cancers 2019, 11, 774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanabe, K.; Kitagawa, K.; Kojima, N.; Iijima, S. Multifucosylated Alpha-1-acid Glycoprotein as a Novel Marker for Hepatocellular Carcinoma. J. Proteome Res. 2016, 15, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Mikami, M.; Tanabe, K.; Matsuo, K.; Miyazaki, Y.; Miyazawa, M.; Hayashi, M.; Asai, S.; Ikeda, M.; Shida, M.; Hirasawa, T.; et al. Fully-sialylated alpha-chain of complement 4-binding protein: Diagnostic utility for ovarian clear cell carcinoma. Gynecol. Oncol. 2015, 139, 520–528. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, K.; Tanabe, K.; Ikeda, M.; Shibata, T.; Kajiwara, H.; Miyazawa, M.; Miyazawa, M.; Hayashi, M.; Shida, M.; Hirasawa, T.; et al. Fully sialylated alpha-chain of complement 4-binding protein (A2160): A novel prognostic marker for epithelial ovarian carcinoma. Arch. Gynecol. Obs. 2018, 297, 749–756. [Google Scholar] [CrossRef]
- Tanabe, K.; Matsuo, K.; Miyazawa, M.; Hayashi, M.; Ikeda, M.; Shida, M.; Hirasawa, T.; Sho, R.; Mikami, M. UPLC-MS/MS based diagnostics for epithelial ovarian cancer using fully sialylated C4-binding protein. Biomed. Chromatogr. BMC 2018, 32, e4180. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, M.; Matsuo, K.; Tanabe, K.; Ikeda, M.; Miyazawa, M.; Yasaka, M.; Machida, H.; Shida, M.; Imanishi, T.; Grubbs, B.H.; et al. Comprehensive Serum Glycopeptide Spectra Analysis (CSGSA): A Potential New Tool for Early Detection of Ovarian Cancer. Cancers 2019, 11, 591. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Chuang, K.V.; DeCarli, C.; Jin, L.W.; Beckett, L.; Keiser, M.J.; Dugger, B.N. Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline. Nat. Commun. 2019, 10, 2173. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Liao, S.; Hu, W. Efficient Single-Stage Pedestrian Detector by Asymptotic Localization Fitting and Multi-Scale Context Encoding. IEEE Trans. Image Process. Publ. IEEE Signal Process. Soc. 2019, 29, 1413–1425. [Google Scholar] [CrossRef]
- Ding, C.; Tao, D. Trunk-Branch Ensemble Convolutional Neural Networks for Video-Based Face Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 1002–1014. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.; Kim, H.Y. Forecasting stock prices with a feature fusion LSTM-CNN model using different representations of the same data. PLoS ONE 2019, 14, e0212320. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef] [Green Version]
- Oh, K.; Chung, Y.C.; Kim, K.W.; Kim, W.S.; Oh, I.S. Classification and Visualization of Alzheimer’s Disease using Volumetric Convolutional Neural Network and Transfer Learning. Sci. Rep. 2019, 9, 18150. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [Green Version]
- Raghu, S.; Sriraam, N.; Temel, Y.; Rao, S.V.; Kubben, P.L. EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Netw. Off. J. Int. Neural Netw. Soc. 2020, 124, 202–212. [Google Scholar] [CrossRef]
- Lee, K.S.; Jung, S.K.; Ryu, J.J.; Shin, S.W.; Choi, J. Evaluation of Transfer Learning with Deep Convolutional Neural Networks for Screening Osteoporosis in Dental Panoramic Radiographs. J. Clin. Med. 2020, 9, 392. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Vans, E.; Shigemizu, D.; Boroevich, K.A.; Tsunoda, T. DeepInsight: A methodology to transform a non-image data to an image for convolution neural network architecture. Sci. Rep. 2019, 9, 11399. [Google Scholar] [CrossRef] [Green Version]
- MathWorks. Alexnet. Available online: https://jp.mathworks.com/help/deeplearning/ref/alexnet.html?lang=en (accessed on 5 August 2020).
- Li, F.-F.; Deng, J.; Russakovsky, O.; Berg, A.; Li, K. ImageNet. Available online: http://www.image-net.org/ (accessed on 19 May 2020).
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the Neural Information Processing Systems Conference, Montreal, QC, Canada, 8–13 December 2014; pp. 2672–2680. [Google Scholar]
- Marouf, M.; Machart, P.; Bansal, V.; Kilian, C.; Magruder, D.S.; Krebs, C.F.; Bonn, S. Realistic in silico generation and augmentation of single-cell RNA-seq data using generative adversarial networks. Nat. Commun. 2020, 11, 166. [Google Scholar] [CrossRef] [Green Version]
- Prat, J. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynaecol. Obstet. Off. Organ Int. Fed. Gynaecol. Obstet. 2014, 124, 1–5. [Google Scholar] [CrossRef]
- Shinozaki, E.; Tanabe, K.; Akiyoshi, T.; Tsuchida, T.; Miyazaki, Y.; Kojima, N.; Igarashi, M.; Ueno, M.; Suenaga, M.; Mizunuma, N.; et al. Serum leucine-rich alpha-2-glycoprotein-1 with fucosylated triantennary N-glycan: A novel colorectal cancer marker. BMC Cancer 2018, 18, 406. [Google Scholar] [CrossRef] [PubMed]





| Number | Layer Name | Role | Description |
|---|---|---|---|
| 1 | data | Image Input | 227 × 227 × 3 images with “zero centering” normalization |
| 2 | conv1 | Convolution | 96 × 11, 11 × 3 convolutions with stride [4 4] and padding [0 0 0 0] |
| 3 | relu1 | ReLU | ReLU |
| 4 | norm1 | Cross Channel Normalization | cross channel normalization with 5 channels per element |
| 5 | pool1 | Max Pooling | 3 × 3 max pooling with stride [2 2] and padding [0 0 0 0] |
| 6 | conv2 | Grouped Convolution | 2 groups of 128 (5 × 5 × 48) convolutions with stride [1 1] and padding [2 2 2 2] |
| 7 | relu2′ | ReLU | ReLU |
| 8 | norm2 | Cross Channel Normalization | cross channel normalization with 5 channels per element |
| 9 | pool2 | Max Pooling | 3 × 3 max pooling with stride [2 2] and padding [0 0 0 0] |
| 10 | conv3 | Convolution | 384 3 × 3 × 256 convolutions with stride [1 1] and padding [1 1 1 1] |
| 11 | relu3 | ReLU | ReLU |
| 12 | conv4 | Grouped Convolution | 2 groups of 192 3 × 3 × 192 convolutions with stride [1 1] and padding [1 1 1 1] |
| 13 | relu4 | ReLU | ReLU |
| 14 | conv5 | Grouped Convolution | 2 groups of 128 3 × 3 × 192 convolutions with stride [1 1] and padding [1 1 1 1] |
| 15 | relu5 | ReLU | ReLU |
| 16 | pool5 | Max Pooling | 3 × 3 max pooling with stride [2 2] and padding [0 0 0 0] |
| 17 | fc6 | Fully Connected | 4096 fully connected layer |
| 18 | relu6 | ReLU | ReLU |
| 19 | drop6 | Dropout | 50% dropout |
| 20 | fc7 | Fully Connected | 4096 fully connected layer |
| 21 | relu7 | ReLU | ReLU |
| 22 | drop7 | Dropout | 50% dropout |
| 23 | fc8 | Fully Connected | 1000 fully connected layer |
| 24 | prob | Softmax | softmax |
| 25 | output | Classification Output | crossentropyex with “EOC” and “Non-EOC” |
| CSGSA-AI (Test) (Cut Off = 0.5) | ||||||
|---|---|---|---|---|---|---|
| Condition | Total | PPV and NPV | ||||
| EOC Stage1 | Non | |||||
| CSGSA-AI | Pos | 31 | 4 | 35 | PPV | 89% |
| Neg | 8 | 98 | 106 | NPV | 92% | |
| Total | 39 | 102 | 141 | |||
| Sensitivity and Specificity | Sens | Spec | Accuracy | |||
| 79% | 96% | 91% | ||||
| Class | Cases | Age | |
|---|---|---|---|
| EOC Stage I (n = 97) | Clear cell carcinoma | 41 | 54.4 ± 12.9 |
| Mucinous carcinoma | 14 | ||
| Endometrioid carcinoma | 28 | ||
| Serous adenocarcinoma | 13 | ||
| Unclassified | 1 | ||
| Non-EOC (n = 254) | Healthy | 220 | 53.8 ± 11.4 |
| Uterine fibroid | 20 | ||
| Ovarian cyst | 14 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanabe, K.; Ikeda, M.; Hayashi, M.; Matsuo, K.; Yasaka, M.; Machida, H.; Shida, M.; Katahira, T.; Imanishi, T.; Hirasawa, T.; et al. Comprehensive Serum Glycopeptide Spectra Analysis Combined with Artificial Intelligence (CSGSA-AI) to Diagnose Early-Stage Ovarian Cancer. Cancers 2020, 12, 2373. https://doi.org/10.3390/cancers12092373
Tanabe K, Ikeda M, Hayashi M, Matsuo K, Yasaka M, Machida H, Shida M, Katahira T, Imanishi T, Hirasawa T, et al. Comprehensive Serum Glycopeptide Spectra Analysis Combined with Artificial Intelligence (CSGSA-AI) to Diagnose Early-Stage Ovarian Cancer. Cancers. 2020; 12(9):2373. https://doi.org/10.3390/cancers12092373
Chicago/Turabian StyleTanabe, Kazuhiro, Masae Ikeda, Masaru Hayashi, Koji Matsuo, Miwa Yasaka, Hiroko Machida, Masako Shida, Tomoko Katahira, Tadashi Imanishi, Takeshi Hirasawa, and et al. 2020. "Comprehensive Serum Glycopeptide Spectra Analysis Combined with Artificial Intelligence (CSGSA-AI) to Diagnose Early-Stage Ovarian Cancer" Cancers 12, no. 9: 2373. https://doi.org/10.3390/cancers12092373
APA StyleTanabe, K., Ikeda, M., Hayashi, M., Matsuo, K., Yasaka, M., Machida, H., Shida, M., Katahira, T., Imanishi, T., Hirasawa, T., Sato, K., Yoshida, H., & Mikami, M. (2020). Comprehensive Serum Glycopeptide Spectra Analysis Combined with Artificial Intelligence (CSGSA-AI) to Diagnose Early-Stage Ovarian Cancer. Cancers, 12(9), 2373. https://doi.org/10.3390/cancers12092373