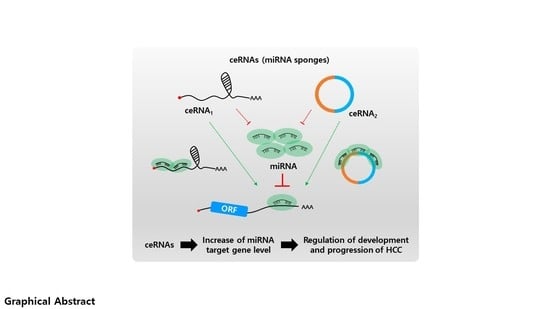
Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Long Non-Coding RNA and microRNA Networks in HCC
2.1. Overexpression of lncRNAs Promotes HCC Proliferation and Metastasis via Sponging of Tumor Suppressing miRNAs
2.2. Tumor Suppressor lncRNAs Inhibit HCC Tumorigenesis by Sponging Onco-miRNAs
3. Circular RNA and microRNA Networks in HCC
3.1. Overexpression of Circrnas Induces HCC Progression by Sponging Tumor-Suppressive miRNAs
3.2. Tumor Suppressor Circrnas Inhibit HCC Tumorigenesis by Sponging Onco-miRNAs
4. Clinical Application of lncRNAs and CircRNAs as Novel Biomarkers in HCC
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Parikh, S.; Hyman, D. Hepatocellular cancer: A guide for the internist. Am. J. Med. 2007, 120, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Kao, J.H. Hepatitis B virus-related hepatocellular carcinoma: Epidemiology and pathogenic role of viral factors. J. Chin. Med. Assoc. 2007, 70, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Turati, F.; Galeone, C.; Rota, M.; Pelucchi, C.; Negri, E.; Bagnardi, V.; Corrao, G.; Boffetta, P.; La Vecchia, C. Alcohol and liver cancer: A systematic review and meta-analysis of prospective studies. Ann. Oncol. 2014, 25, 1526–1535. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.W.; Lin, C.L.; Liu, C.J.; Yang, S.H.; Tseng, Y.L.; Wu, C.F. Influence of Metabolic Risk Factors on Risk of Hepatocellular Carcinoma and Liver-Related Death in Men With Chronic Hepatitis B: A Large Cohort Study. Gastroenterology 2017, 153, 1006–1017. [Google Scholar] [CrossRef]
- Torres, H.A.; Shigle, T.L.; Hammoudi, N.; Link, J.T.; Samaniego, F.; Kaseb, A.; Mallet, V. The oncologic burden of hepatitis C virus infection: A clinical perspective. CA Cancer J. Clin. 2017, 67, 411–431. [Google Scholar] [CrossRef]
- Nakagawa, S.; Wei, L.; Song, W.M.; Higashi, T.; Ghoshal, S.; Kim, R.S.; Bian, C.B.; Yamada, S.; Sun, X.; Venkatesh, A.; et al. Molecular Liver Cancer Prevention in Cirrhosis by Organ Transcriptome Analysis and Lysophosphatidic Acid Pathway Inhibition. Cancer Cell 2016, 30, 879–890. [Google Scholar] [CrossRef]
- Parikh, N.D.; Singal, A.G.; Hutton, D.W. Cost effectiveness of regorafenib as second-line therapy for patients with advanced hepatocellular carcinoma. Cancer 2017, 123, 3725–3731. [Google Scholar] [CrossRef] [Green Version]
- Goossens, N.; Sun, X.; Hoshida, Y. Molecular classification of hepatocellular carcinoma: Potential therapeutic implications. Hepat. Oncol. 2015, 2, 371–379. [Google Scholar] [CrossRef]
- Cao, Z.; Fan-Minogue, H.; Bellovin, D.I.; Yevtodiyenko, A.; Arzeno, J.; Yang, Q.; Gambhir, S.S.; Felsher, D.W. MYC phosphorylation, activation, and tumorigenic potential in hepatocellular carcinoma are regulated by HMG-CoA reductase. Cancer Res. 2011, 71, 2286–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.M.; Liu, Y.L.; Lin, Y.C.; Shun, C.T.; Wu, M.S.; Chen, C.C. Inhibition of autophagy enhances anticancer effects of atorvastatin in digestive malignancies. Cancer Res. 2010, 70, 7699–7709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutter, A.P.; Maaser, K.; Hopfner, M.; Huether, A.; Schuppan, D.; Scherubl, H. Cell cycle arrest and apoptosis induction in hepatocellular carcinoma cells by HMG-CoA reductase inhibitors. Synergistic antiproliferative action with ligands of the peripheral benzodiazepine receptor. J. Hepatol. 2005, 43, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Fuchs, B.C.; Tanabe, K.K. Prevention of hepatocellular carcinoma: Potential targets, experimental models, and clinical challenges. Curr. Cancer Drug Targets 2012, 12, 1129–1159. [Google Scholar]
- Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Wong, C.M.; Tsang, F.H.; Ng, I.O. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, W.; Shen, W.; Xia, M.; Chen, C.; Xiang, D.; Ning, B.; Cui, X.; Li, H.; Li, X.; et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 2016, 64, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Xu, Q.G.; Wang, Z.G.; Yang, Y.; Zhang, L.; Ma, J.Z.; Sun, S.H.; Yang, F.; Zhou, W.P. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018, 68, 1214–1227. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Toth, K.F.; Aravin, A.A. piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 2017, 33, 882–894. [Google Scholar] [CrossRef] [Green Version]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA-target interactions—Beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef]
- Bassett, A.R.; Azzam, G.; Wheatley, L.; Tibbit, C.; Rajakumar, T.; McGowan, S.; Stanger, N.; Ewels, P.A.; Taylor, S.; Ponting, C.P.; et al. Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat. Commun. 2014, 5, 4640. [Google Scholar] [CrossRef]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Han, T.S.; Hur, K.; Xu, G.; Choi, B.; Okugawa, Y.; Toiyama, Y.; Oshima, H.; Oshima, M.; Lee, H.J.; Kim, V.N.; et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2015, 64, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Han, T.S.; Voon, D.C.; Oshima, H.; Nakayama, M.; Echizen, K.; Sakai, E.; Yong, Z.W.E.; Murakami, K.; Yu, L.; Minamoto, T.; et al. Interleukin 1 Up-regulates MicroRNA 135b to Promote Inflammation-Associated Gastric Carcinogenesis in Mice. Gastroenterology 2019, 156, 1140–1155. [Google Scholar] [CrossRef]
- Hwang, J.S.; Jeong, E.J.; Choi, J.; Lee, Y.J.; Jung, E.; Kim, S.K.; Min, J.K.; Han, T.S.; Kim, J.S. MicroRNA-1258 Inhibits the Proliferation and Migration of Human Colorectal Cancer Cells through Suppressing CKS1B Expression. Genes 2019, 10, 912. [Google Scholar] [CrossRef] [Green Version]
- Law, P.T.; Qin, H.; Ching, A.K.; Lai, K.P.; Co, N.N.; He, M.; Lung, R.W.; Chan, A.W.; Chan, T.F.; Wong, N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J. Hepatol. 2013, 58, 1165–1173. [Google Scholar] [CrossRef]
- Sandbothe, M.; Buurman, R.; Reich, N.; Greiwe, L.; Vajen, B.; Gurlevik, E.; Schaffer, V.; Eilers, M.; Kuhnel, F.; Vaquero, A.; et al. The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J. Hepatol. 2017, 66, 1012–1021. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Han, T.S.; Ban, H.S.; Hur, K.; Cho, H.S. The Epigenetic Regulation of HCC Metastasis. Int. J. Mol. Sci. 2018, 19, 3978. [Google Scholar] [CrossRef] [Green Version]
- Bae, H.J.; Jung, K.H.; Eun, J.W.; Shen, Q.; Kim, H.S.; Park, S.J.; Shin, W.C.; Yang, H.D.; Park, W.S.; Lee, J.Y.; et al. MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J. Hepatol. 2015, 63, 408–419. [Google Scholar] [CrossRef]
- Liu, S.; Guo, W.; Shi, J.; Li, N.; Yu, X.; Xue, J.; Fu, X.; Chu, K.; Lu, C.; Zhao, J.; et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J. Hepatol. 2012, 56, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.E.; Nitsch, R.; Wulczyn, F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008, 10, 987–993. [Google Scholar] [CrossRef]
- Sayed, D.; Rane, S.; Lypowy, J.; He, M.; Chen, I.Y.; Vashistha, H.; Yan, L.; Malhotra, A.; Vatner, D.; Abdellatif, M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell 2008, 19, 3272–3282. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Ono, K.; Nishi, H.; Iwanaga, Y.; Nagao, K.; Kinoshita, M.; Kuwabara, Y.; Takanabe, R.; Hasegawa, K.; Kita, T.; et al. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem. Biophys. Res. Commun. 2009, 389, 315–320. [Google Scholar] [CrossRef] [Green Version]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Timmermans, M.C. Target mimics modulate miRNAs. Nat. Genet. 2007, 39, 935–936. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Han, T.; Li, Y.; Sun, J.; Zhang, S.; Liu, Y.; Shan, B.; Zheng, D.; Shi, J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017, 31, 893–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Huang, F.; Lei, Z.; Luo, H. LncRNA SNHG11 Promotes Proliferation, Migration, Apoptosis, and Autophagy by Regulating hsa-miR-184/AGO2 in HCC. Onco Targets Ther. 2020, 13, 413–421. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Yang, S.B.; Xu, F.F.; Zhang, J.H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J. Exp. Clin. Cancer Res. 2015, 34, 18. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Peng, F.; Ning, Y.; Jiang, P.; Peng, J.; Ding, X.; Zhang, J.; Jiang, T.; Xiang, S. SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J. Cell. Biochem. 2020, 121, 2543–2558. [Google Scholar] [CrossRef]
- Huang, Y.; Xiang, B.; Liu, Y.; Wang, Y.; Kan, H. LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer Lett. 2018, 437, 56–66. [Google Scholar] [CrossRef]
- Gong, J.; Wang, J.; Liu, T.; Hu, J.; Zheng, J. lncRNA FEZF1AS1 contributes to cell proliferation, migration and invasion by sponging miR4443 in hepatocellular carcinoma. Mol. Med. Rep. 2018, 18, 5614–5620. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.Q.; Li, L.; Lu, C.; Liu, J.; Chen, Y.; Wu, H. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J. Cell. Physiol. 2019, 234, 5153–5162. [Google Scholar] [CrossRef]
- Pan, Y.; Tong, S.; Cui, R.; Fan, J.; Liu, C.; Lin, Y.; Tang, J.; Xie, H.; Lin, P.; Zheng, T.; et al. Long Non-Coding MALAT1 Functions as a Competing Endogenous RNA to Regulate Vimentin Expression by Sponging miR-30a-5p in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2018, 50, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Miao, R.; Liu, S.; Wan, Y.; Zhang, S.; Deng, Y.; Bi, J.; Qu, K.; Zhang, J.; Liu, C. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget 2017, 8, 28683–28695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.; Zhang, J.; Zhou, C.; Qiu, G.; Wang, G.; Wang, S.; Chang, X.; Li, X.; Fan, L. Long non-coding RNA FOXD2-AS1 plays an oncogenic role in hepatocellular carcinoma by targeting miR206. Oncol. Rep. 2018, 40, 3625–3634. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Han, Y.; Zhang, Y.; Zhou, Y.; Ye, L. lncRNA TINCR sponges miR-214-5p to upregulate ROCK1 in hepatocellular carcinoma. BMC Med. Genet. 2020, 21, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, W.; Dai, J.L.; Tang, M.H.; Ye, M.S.; Fang, S. lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/ histone deacetylase 2 axis. World J. Gastroenterol. 2019, 25, 5789–5799. [Google Scholar] [CrossRef]
- Dong, J.; Teng, F.; Guo, W.; Yang, J.; Ding, G.; Fu, Z. lncRNA SNHG8 Promotes the Tumorigenesis and Metastasis by Sponging miR-149-5p and Predicts Tumor Recurrence in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2018, 51, 2262–2274. [Google Scholar] [CrossRef]
- Zhang, K.; Zhao, Z.; Yu, J.; Chen, W.; Xu, Q.; Chen, L. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 6045–6056. [Google Scholar] [CrossRef]
- Li, C.; Lu, L.; Feng, B.; Zhang, K.; Han, S.; Hou, D.; Chen, L.; Chu, X.; Wang, R. The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci. Rep. 2017, 7, 4637. [Google Scholar] [CrossRef]
- Tu, J.; Zhao, Z.; Xu, M.; Chen, M.; Weng, Q.; Ji, J. LINC00460 promotes hepatocellular carcinoma development through sponging miR-485-5p to up-regulate PAK1. Biomed. Pharmacother. 2019, 118, 109213. [Google Scholar] [CrossRef]
- Gao, J.; Yin, X.; Yu, X.; Dai, C.; Zhou, F. Long noncoding RNA LINC00488 functions as a ceRNA to regulate hepatocellular carcinoma cell growth and angiogenesis through miR-330-5. Dig. Liver Dis. 2019, 51, 1050–1059. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, L.; Wang, L.; Liu, Z.; Li, Q.; Yao, B.; Wang, C.; Chen, T.; Tu, K.; Liu, Q. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/beta-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 851. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Chen, X.; Hou, Y.; Kang, G.; Li, Q.; Xu, Y. LncRNA DBH-AS1 facilitates the tumorigenesis of hepatocellular carcinoma by targeting miR-138 via FAK/Src/ERK pathway. Biomed. Pharmacother. 2018, 107, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Kong, Y.; Gao, Z.; Liu, Y.; Zhu, P.; Yu, Z. LncRNA TUG1 interacting with miR-144 contributes to proliferation, migration and tumorigenesis through activating the JAK2/STAT3 pathway in hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 2018, 101, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Ma, W.; Hong, Z.; Wu, L.; Chen, X.; Yuan, Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 11. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.; Zhang, T.; Zhang, D.; Xie, L.; Zou, X.; Lei, L.; Wu, D.; Liu, L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene 2017, 36, 1112–1122. [Google Scholar] [CrossRef]
- Wang, F.; Ying, H.Q.; He, B.S.; Pan, Y.Q.; Deng, Q.W.; Sun, H.L.; Chen, J.; Liu, X.; Wang, S.K. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 2015, 6, 7899–7917. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhou, Y.; Yang, L.; Ma, Y.; Peng, X.; Yang, S.; Li, H.; Liu, J. LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J. Cell. Physiol. 2020, 235, 3402–3413. [Google Scholar] [CrossRef]
- Guo, J.; Ma, Y.; Peng, X.; Jin, H.; Liu, J. LncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging miR-181 in hepatocellular carcinoma. J. Cell. Biochem. 2019, 120, 17975–17983. [Google Scholar] [CrossRef]
- Yang, L.; Peng, X.; Jin, H.; Liu, J. Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene 2019, 697, 94–102. [Google Scholar] [CrossRef]
- Zhao, L.; Hu, K.; Cao, J.; Wang, P.; Li, J.; Zeng, K.; He, X.; Tu, P.F.; Tong, T.; Han, L. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging 2019, 11, 7098–7122. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.; Sun, H.; Pan, Y.; Wu, M.; Zhang, J. Deregulation of miR-520d-3p promotes hepatocellular carcinoma development via lncRNA MIAT regulation and EPHA2 signaling activation. Biomed. Pharmacother. 2019, 109, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ke, S.; Li, M.; Lin, C.; Liu, X.; Pan, Q. Downregulation of LncRNA GAS5 promotes liver cancer proliferation and drug resistance by decreasing PTEN expression. Mol. Genet. Genom. 2020, 295, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zha, G.; Wu, Y.; Cai, W.; Ao, J. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018, 11, 8855–8863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhu, Z.; Huang, S.; Zhao, Q.; Huang, C.; Tang, Y.; Sun, C.; Zhang, Z.; Wang, L.; Chen, H.; et al. lncRNA XIST regulates proliferation and migration of hepatocellular carcinoma cells by acting as miR-497-5p molecular sponge and targeting PDCD4. Cancer Cell. Int. 2019, 19, 198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.G.; Wang, T.; Shi, M.; Zhai, B. Long noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma development by sponging miR-301a-5p and targeting FOXL1. J. Exp. Clin. Cancer Res. 2019, 38, 153. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.G.; Liu, J.; Shi, M.; Chen, F.X. LncRNA DGCR5 represses the development of hepatocellular carcinoma by targeting the miR-346/KLF14 axis. J. Cell. Physiol. 2018, 234, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Tang, Z.; Chen, K.; Liu, Y.; Yu, G.; Chen, Q.; Dang, H.; Chen, F.; Ling, J.; Zhu, L.; et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J. Exp. Clin. Cancer Res. 2018, 37, 214. [Google Scholar] [CrossRef]
- Hu, B.; Cai, H.; Zheng, R.; Yang, S.; Zhou, Z.; Tu, J. Long non-coding RNA 657 suppresses hepatocellular carcinoma cell growth by acting as a molecular sponge of miR-106a-5p to regulate PTEN expression. Int. J. Biochem. Cell Biol. 2017, 92, 34–42. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Z.; Yao, B.; Dou, C.; Xu, M.; Xue, Y.; Ding, L.; Jia, Y.; Zhang, H.; Li, Q.; et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumor Biol. 2016, 37, 11429–11441. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Shi, H.; Yuan, J.; Qiao, L.; Dong, L.; Wang, Y. Downregulation of circular RNA circPVT1 restricts cell growth of hepatocellular carcinoma through downregulation of Sirtuin 7 via microRNA-3666. Clin. Exp. Pharmacol. Physiol. 2020. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Zheng, Q.; Bao, C.; He, J.; Chen, B.; Lyu, D.; Zheng, B.; Xu, Y.; Long, Z.; et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017, 388, 208–219. [Google Scholar] [CrossRef]
- Wang, Z.; Su, M.; Xiang, B.; Zhao, K.; Qin, B. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem. Biophys. Res. Commun. 2019, 512, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Zhao, Y.; Lim, G.; Lin, H.; Zhang, X.; Zhang, X. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed. Pharmacother. 2019, 111, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, G.; Sun, Y.; Gao, Y.; Ouyang, X.; Chang, C.; Gong, L.; Yeh, S. Targeting the estrogen receptor alpha (ERalpha)-mediated circ-SMG1.72/miR-141-3p/Gelsolin signaling to better suppress the HCC cell invasion. Oncogene 2020, 39, 2493–2508. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Huang, Z.L.; Xu, Y.H.; Zheng, Q.; Chen, Z.; Song, W.; Zhou, J.; Tang, Z.Y.; Huang, X.Y. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 2017, 7, 5428. [Google Scholar] [CrossRef]
- Wu, J.; Liu, S.; Xiang, Y.; Qu, X.; Xie, Y.; Zhang, X. Bioinformatic Analysis of Circular RNA-Associated ceRNA Network Associated with Hepatocellular Carcinoma. BioMed Res. Int. 2019, 2019, 8308694. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, X.; Liang, C.; Ling, Y.; Yang, X.; Ye, X.; Zhang, H.; Yang, P.; Cui, X.; Ren, Y.; et al. A Noncoding Regulatory RNAs Network Driven by Circ-CDYL Acts Specifically in the Early Stages Hepatocellular Carcinoma. Hepatology 2020, 71, 130–147. [Google Scholar] [CrossRef]
- Qi, S.X.; Sun, H.; Liu, H.; Yu, J.; Jiang, Z.Y.; Yan, P. Role and mechanism of circ-PRKCI in hepatocellular carcinoma. World J. Gastroenterol. 2019, 25, 1964–1974. [Google Scholar] [CrossRef]
- Xie, B.; Zhao, Z.; Liu, Q.; Wang, X.; Ma, Z.; Li, H. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene 2019, 683, 253–261. [Google Scholar] [CrossRef]
- Liu, H.; Xue, L.; Song, C.; Liu, F.; Jiang, T.; Yang, X. Overexpression of circular RNA circ_001569 indicates poor prognosis in hepatocellular carcinoma and promotes cell growth and metastasis by sponging miR-411-5p and miR-432-5p. Biochem. Biophys. Res. Commun. 2018, 503, 2659–2665. [Google Scholar] [CrossRef]
- Li, M.F.; Li, Y.H.; He, Y.H.; Wang, Q.; Zhang, Y.; Li, X.F.; Meng, X.M.; Huang, C.; Li, J. Emerging roles of hsa_circ_0005075 targeting miR-431 in the progress of HCC. Biomed. Pharmacother. 2018, 99, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Bai, N.; Peng, E.; Qiu, X.; Lyu, N.; Zhang, Z.; Tao, Y.; Li, X.; Wang, Z. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 2018, 37, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Shi, Y.; Liu, M.; Sun, J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yu, C.; Wu, M.; Wu, X.; Wan, L.; Zhu, X. MicroRNA-191 promotes hepatocellular carcinoma cell proliferation by has_circ_0000204/miR-191/KLF6 axis. Cell Prolif. 2019, 52, e12635. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhao, Y.; Wang, Y.; Jin, C. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed. Pharmacother. 2019, 116, 108932. [Google Scholar] [CrossRef]
- Xu, L.; Feng, X.; Hao, X.; Wang, P.; Zhang, Y.; Zheng, X.; Li, L.; Ren, S.; Zhang, M.; Xu, M. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J. Exp. Clin. Cancer Res. 2019, 38, 98. [Google Scholar] [CrossRef] [Green Version]
- Qiu, L.; Huang, Y.; Li, Z.; Dong, X.; Chen, G.; Xu, H.; Zeng, Y.; Cai, Z.; Liu, X.; Liu, J. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol. Oncol. 2019, 13, 441–455. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Chen, Q.; Yao, T.; Li, T.; Ying, S.; Hu, Y.; Guo, J. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 2017, 8, 43878–43888. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhang, Y.; Wu, X.; Zhang, C.; Li, G. Diagnostic Value of lncRNAs as Biomarker in Hepatocellular Carcinoma: An Updated Meta-Analysis. Can. J. Gastroenterol. Hepatol. 2018, 2018, 8410195. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Shi, H.; Wang, X.; Wang, B.; Qu, Q.; Geng, H.; Sun, H. Identification of diagnostic long noncoding RNA biomarkers in patients with hepatocellular carcinoma. Mol. Med. Rep. 2019, 20, 1121–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.; Li, Y.; Li, M.; Wang, L. Long noncoding RNA GAS5 inhibits metastasis by targeting miR-182/ANGPTL1 in hepatocellular carcinoma. Am. J. Cancer Res. 2019, 9, 108–121. [Google Scholar]
- Han, M.H.; Lee, J.H.; Kim, G.; Lee, E.; Lee, Y.R.; Jang, S.Y.; Lee, H.W.; Chun, J.M.; Han, Y.S.; Yoon, J.S.; et al. Expression of the Long Noncoding RNA GAS5 Correlates with Liver Fibrosis in Patients with Nonalcoholic Fatty Liver Disease. Genes 2020, 11, 545. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ye, H.; Huang, G.; Luo, F.; Liu, Y.; Liu, Y.; Yang, X.; Shen, J.; Liu, Q.; Zhang, J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016, 37, 2691–2702. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.Q.; Li, R.J.; Mei, J.Z.; Li, X.H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 4303–4309. [Google Scholar]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, F.; Zhao, M.; Yang, Z.; Li, J.; Zhang, S.; Zhang, W.; Ye, L.; Zhang, X. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J. Biol. Chem. 2017, 292, 15395–15407. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, X.; Tang, J.; Jiang, R.; Zhang, W.; Ji, J.; Sun, B. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2015, 37, 687–696. [Google Scholar] [CrossRef]
- Konishi, H.; Ichikawa, D.; Yamamoto, Y.; Arita, T.; Shoda, K.; Hiramoto, H.; Hamada, J.; Itoh, H.; Fujita, Y.; Komatsu, S.; et al. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci. 2016, 107, 149–154. [Google Scholar] [CrossRef]
- Zheng, Z.K.; Pang, C.; Yang, Y.; Duan, Q.; Zhang, J.; Liu, W.C. Serum long noncoding RNA urothelial carcinoma-associated 1: A novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J. Int. Med. Res. 2018, 46, 348–356. [Google Scholar] [CrossRef] [Green Version]
- Roberts, L.R. Current Status of the GALAD and BALAD Biomarker Models for Hepatocellular Carcinoma. Gastroenterol. Hepatol. 2019, 15, 672–675. [Google Scholar]
- Best, J.; Bechmann, L.P.; Sowa, J.P.; Sydor, S.; Dechene, A.; Pflanz, K.; Bedreli, S.; Schotten, C.; Geier, A.; Berg, T.; et al. GALAD Score Detects Early Hepatocellular Carcinoma in an International Cohort of Patients With Nonalcoholic Steatohepatitis. Clin. Gastroenterol. Hepatol. 2020, 18, 728–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, X.; Li, G.; Liu, H.; Li, T.; Liu, J.; Zhao, Q.; Wang, C. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine 2016, 95, e3811. [Google Scholar] [CrossRef]
- Jin, Y.D.; Ren, Y.R.; Gao, Y.X.; Zhang, L.; Ding, Z. Hsa_circ_0005075 predicts a poor prognosis and acts as an oncogene in colorectal cancer via activating Wnt/beta-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3311–3319. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, C.; Lin, J.; Song, X.; Wang, H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag. Res. 2018, 10, 1275–1283. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Yao, T.; Chen, Q.; Mo, X.; Hu, Y.; Guo, J. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget 2017, 8, 58405–58416. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Gu, B.; Yao, G.; Li, P.; Wang, K. Circular RNA Expression Profiles and the Pro-tumorigenic Function of CircRNA_10156 in Hepatitis B Virus-Related Liver Cancer. Int. J. Med. Sci. 2020, 17, 1351–1365. [Google Scholar] [CrossRef]
- Memczak, S.; Papavasileiou, P.; Peters, O.; Rajewsky, N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS ONE 2015, 10, e0141214. [Google Scholar] [CrossRef]
- Mehinovic, L.; Islamagic, E.; Husic-Selimovic, A.; Kurtovic-Kozaric, A.; Vukobrat-Bijedic, Z.; Suljevic, D. Evaluation of Diagnostic Efficiency of Alpha-Fetoprotein in Patients with Liver Cirrhosis and Hepatocellular Carcinoma: Single-Center Experience. Open Access Maced. J. Med. Sci. 2018, 6, 1668–1673. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Chen, C.; Yang, Q.; Xue, J.; Chen, X.; Sun, B.; Luo, F.; Liu, X.; Xiao, T.; Xu, H.; et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018, 9, 454. [Google Scholar] [CrossRef]
- Wang, G.; Liu, W.; Zou, Y.; Wang, G.; Deng, Y.; Luo, J.; Zhang, Y.; Li, H.; Zhang, Q.; Yang, Y.; et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine 2019, 40, 432–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Deng, T.; Ge, S.; Liu, Y.; Bai, M.; Zhu, K.; Fan, Q.; Li, J.; Ning, T.; Tian, F.; et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 2019, 38, 2844–2859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Chen, Z.; Zhang, W.; Cheng, Y.; Zhang, B.; Wu, F.; Wang, Q.; Wang, S.; Rong, D.; Reiter, F.P.; et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: Theoretical basis and therapeutic aspects. Signal Transduct. Target. Ther. 2020, 5, 87. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Chen, L.; Cai, X.; Zhang, Y.; Zhang, J.; Ma, D.; Cai, X.; Fu, T.; Yu, Z.; Yu, F.; et al. Long non-coding RNA TUC338 is functionally involved in sorafenib-sensitized hepatocarcinoma cells by targeting RASAL1. Oncol. Rep. 2017, 37, 273–280. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol. Cancer Res. 2014, 12, 1377–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.Y.; Tang, Y.P.; Liu, J.J.; Liang, R.; Luo, X.L. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J. Cancer 2020, 11, 2993–3001. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef]



| lncRNAs | Target miRNA | Target Genes of miRNA | Function | Reference |
|---|---|---|---|---|
| Overexpression of lncRNAs in HCC | ||||
| SNHG11 | miR-184 | AGO2 | HCC proliferation, migration, autophagy | [55] |
| CCAT1 | let-7 | HMGA2, c-MYC | HCC proliferation, migration | [56] |
| SNHG16 | let-7b-5p | CDC25B | Cell cycle, migration/invasion | [57] |
| HOXA-AS2 | miR-520c-3p | GPC3 | Cell cycle, apoptosis, migration/invasion | [26] |
| CDKN2B-AS1 | let-7c-5p | NAP1L1 | Cell cycle, apoptosis, migration/invasion | [58] |
| FEZF1-AS1 | miR-4443 | HCC proliferation, metastasis | [59] | |
| H19 | miR-326 | TWIST | HCC proliferation, metastasis | [60] |
| MALAT1 | miR-30a-5p miR-146b-5p | Vimentin TRAF6 | HCC proliferation, metastasis | [61] [62] |
| FOXD2-AS1 | miR-206 | ANXA2 | HCC proliferation, metastasis | [63] |
| TINCR | miR-214-5p | ROCK1 | HCC proliferation, metastasis | [64] |
| SNHG15 | miR-490-3P | HDAC2 | HCC proliferation, metastasis | [65] |
| SNHG8 | miR-149-5p | PPM1F | HCC proliferation, metastasis | [66] |
| FLVCR1-AS1 | miR-513c | MET | HCC proliferation, metastasis | [67] |
| ROR | miR-145 | ZEB2 | HCC metastasis | [68] |
| LINC00460 | miR-485-5p | PAK1 | HCC proliferation, angiogenesis | [69] |
| LINC00488 | miR-330-5p | TLN1 | HCC proliferation | [70] |
| DSCR8 | miR-485-5p | FZD7 | Wnt/β-catenin signaling pathway | [71] |
| DBH-AS1 | miR-138 | FAK/Src/ERK pathway | [72] | |
| TUG1 | miR-144 | JAK2/STAT3 pathway | [73] | |
| SNHG12 | miR-199a/b-5p | MLK3 | NF-κB pathway | [74] |
| SNHG6-003 | miR-26a/b | TAK1 | p38 pathway | [75] |
| UCA1 | miR-216b | FGFR1 | FAFR1/ERK signaling pathway | [76] |
| NEAT1 | miR-204 | ATG3 | HCC autophagy process | [77] |
| CCAT1 | miR-181a-5p | ATG7 | HCC autophagy process | [78] |
| PVT1 | miR-365 | ATG3 | HCC autophagy process | [79] |
| MIAT | miR-22-3P miR-520d-3p | SIRT1 EPHA2 | Cellular senescence HCC proliferation, metastasis | [80] [81] |
| Downregulation of lncRNAs in HCC | ||||
| GAS5 | miR-21 | PTEN | Tumor suppressor | [82] |
| SNHG16 | has-miR-93 | Tumor suppressor, 5-FU chemoresistance | [83] | |
| XIST | miR-497-5p | PDCD4 | Tumor suppressor | [84] |
| EPB41L4A-AS2 | miR-301a-5p | FOXL1 | Tumor suppressor | [85] |
| DGCR5 | miR-346 | KLF14 | Tumor suppressor | [86] |
| MIR31HG | miR-575 | ST7L | Tumor suppressor | [87] |
| LINC00657 | miR-106a-5p | PTEN | Tumor suppressor | [88] |
| TUSC7 | miR-10a | EPHA4 | Tumor suppressor | [89] |
| CircRNAs | Target miRNA | Target Genes of miRNA | Function | Reference |
|---|---|---|---|---|
| Overexpression of lncRNAs in HCC | ||||
| circ-PVTa | miR-3666 | SIRTUIN 7 | HCC proliferation | [90] |
| circ-SMG1.72 | miR-141-3p | GELSOLIN | HCC invasion | [94] |
| circ-100338 | miR-141-3p | MTSS1 | hepatitis B-related HCC progression | [95] |
| has-circ-0009910 | miR-1261 | UBE2L3 | HCC progression | [96] |
| circ-CDYL | miR-892a, miR-328-3p | HDGF, HIF1AN | Early stage HCC progression | [97] |
| circ-PRKCI | miRNA-545 | E2F7 | HCC proliferation | [98] |
| has-circ-0078710 | miR-31 | HDAC, CDK2 | HCC progression | [99] |
| circ-001569 | miR-411-5p miR-432-5p | unknown | HCC proliferation, metastasis | [100] |
| has-circ-0005075 | miR-431 | unknown | HCC proliferation, metastasis | [101] |
| circ-FBLIM1 | miR-346 | FBLIM1 | HCC progression | [102] |
| circ-HIPK3 | miR-124 | AQP3 | HCC proliferation, metastasis | [103] |
| Downregulation of lncRNAs in HCC | ||||
| has-circ-0000204 | miR-191 | KLF6 | HCC proliferation | [104] |
| circ-HIAT1 | miR-3171 | PTEN | HCC proliferation | [105] |
| circ-SETD3 (has-circ-0000567) | miR-421 | MAPK14 | HCC proliferation | [106] |
| circ-ADAMTS13 | miR-484 | unknown | HCC proliferation | [107] |
| circ-MTO1 | miR-9 | P21 | HCC progression | [108] |
| has-circ-0005986 | miR-129-5p | NOTCH1 | HCC biomarker | [109] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, T.-S.; Hur, K.; Cho, H.-S.; Ban, H.S. Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers 2020, 12, 2622. https://doi.org/10.3390/cancers12092622
Han T-S, Hur K, Cho H-S, Ban HS. Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers. 2020; 12(9):2622. https://doi.org/10.3390/cancers12092622
Chicago/Turabian StyleHan, Tae-Su, Keun Hur, Hyun-Soo Cho, and Hyun Seung Ban. 2020. "Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma" Cancers 12, no. 9: 2622. https://doi.org/10.3390/cancers12092622
APA StyleHan, T. -S., Hur, K., Cho, H. -S., & Ban, H. S. (2020). Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers, 12(9), 2622. https://doi.org/10.3390/cancers12092622