Glioblastoma Tissue Slice Tandem-Cultures for Quantitative Evaluation of Inhibitory Effects on Invasion and Growth
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
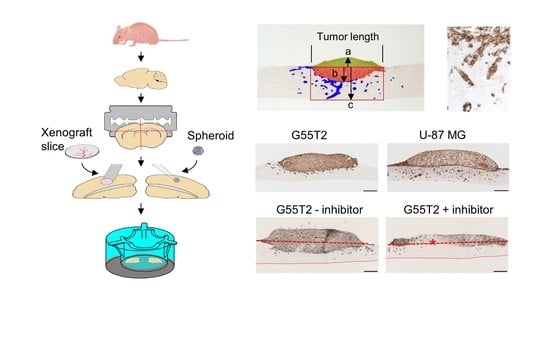
2.1. Tissue Slice Preparation and Cultivation
2.2. Characterization of Tumor Growth and Invasion/Penetration
2.3. Quantitation of Space Occupying Tumor Growth and Tumor Cell Invasion
2.4. Further Characterization of Tissue Slice Xenograft Tandem-Culture Properties in Comparison to Orthotopic Xenografts
2.5. Validation and Testing of the Quantitation of Tumor Invasion Index and Space Occupying Growth Index
2.6. Pim1 Inhibitor Therapy Studies in Tissue Slice Tandem-Cultures
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Cell Culture and Spheroid Generation
4.3. Tumor Cell Xenografting and Tissue Slice Preparation
4.4. Tissue Slice Co-Culture Cultivation and Treatment
4.5. Immunohistochemical Analysis
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathologica 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Claes, A.; Idema, A.J.; Wesseling, P. Diffuse glioma growth: A guerilla war. Acta Neuropathologica 2007, 114, 443–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Nalbantoglu, J. Ring cell migration assay identifies distinct effects of extracellular matrix proteins on cancer cell migration. BMC Res. Notes 2014, 7, 183. [Google Scholar] [CrossRef] [Green Version]
- Nakada, M.; Nambu, E.; Furuyama, N.; Yoshida, Y.; Takino, T.; Hayashi, Y.; Sato, H.; Sai, Y.; Tsuji, T.; Miyamoto, K.I.; et al. Integrin alpha3 is overexpressed in glioma stem-like cells and promotes invasion. Br. J. Cancer 2013, 108, 2516–2524. [Google Scholar] [CrossRef]
- Madsen, C.D.; Hooper, S.; Tozluoglu, M.; Bruckbauer, A.; Fletcher, G.; Erler, J.T.; Bates, P.A.; Thompson, B.; Sahai, E. STRIPAK components determine mode of cancer cell migration and metastasis. Nat. Cell Biol. 2015, 17, 68–80. [Google Scholar] [CrossRef] [Green Version]
- Khatau, S.B.; Bloom, R.J.; Bajpai, S.; Razafsky, D.; Zang, S.; Giri, A.; Wu, P.H.; Marchand, J.; Celedon, A.; Hale, C.M.; et al. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci. Rep. 2012, 2, 488. [Google Scholar] [CrossRef] [Green Version]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef]
- Hattermann, K.; Held-Feindt, J.; Mentlein, R. Spheroid confrontation assay: A simple method to monitor the three-dimensional migration of different cell types in vitro. Annals Anatomy—Anatomischer Anzeigerm 2011, 193, 181–184. [Google Scholar] [CrossRef]
- Humpel, C. Organotypic brain slice cultures: A review. Neuroscience 2015, 305, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Merz, F.; Gaunitz, F.; Dehghani, F.; Renner, C.; Meixensberger, J.; Gutenberg, A.; Giese, A.; Schopow, K.; Hellwig, C.; Schafer, M.; et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro-Oncology 2013, 15, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Merz, L.; Hobel, S.; Kallendrusch, S.; Ewe, A.; Bechmann, I.; Franke, H.; Merz, F.; Aigner, A. Tumor tissue slice cultures as a platform for analyzing tissue-penetration and biological activities of nanoparticles. Eur. J. Pharm. Biopharm. 2017, 112, 45–50. [Google Scholar] [CrossRef]
- Sonnichsen, R.; Hennig, L.; Blaschke, V.; Winter, K.; Korfer, J.; Hahnel, S.; Monecke, A.; Wittekind, C.; Jansen-Winkeln, B.; Thieme, R.; et al. Individual Susceptibility Analysis Using Patient-derived Slice Cultures of Colorectal Carcinoma. Clin. Colorectal Cancer 2018, 17, e189–e199. [Google Scholar] [CrossRef] [PubMed]
- Haehnel, S.; Reiche, K.; Loeffler, D.; Horn, A.; Blumert, C.; Puppel, S.H.; Kaiser, N.; Rapp, F.; Rade, M.; Horn, F.; et al. Deep sequencing and automated histochemistry of human tissue slice cultures improve their usability as preclinical model for cancer research. Sci. Rep. 2019, 9, 19961. [Google Scholar] [CrossRef] [PubMed]
- Gritsenko, P.; Leenders, W.; Friedl, P. Recapitulating in vivo-like plasticity of glioma cell invasion along blood vessels and in astrocyte-rich stroma. Histochem. Cell Biol. 2017, 148, 395–406. [Google Scholar] [CrossRef] [Green Version]
- Aaberg-Jessen, C.; Norregaard, A.; Christensen, K.; Pedersen, C.B.; Andersen, C.; Kristensen, B.W. Invasion of primary glioma- and cell line-derived spheroids implanted into corticostriatal slice cultures. Int. J. Clin. Exp. Pathol. 2013, 6, 546–560. [Google Scholar]
- Carro, M.S.; Lim, W.K.; Alvarez, M.J.; Bollo, R.J.; Zhao, X.; Snyder, E.Y.; Sulman, E.P.; Anne, S.L.; Doetsch, F.; Colman, H.; et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature 2010, 463, 318–325. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Priester, M.; Copanaki, E.; Vafaizadeh, V.; Hensel, S.; Bernreuther, C.; Glatzel, M.; Seifert, V.; Groner, B.; Kogel, D.; Weissenberger, J. STAT3 silencing inhibits glioma single cell infiltration and tumor growth. Neuro-Oncology 2013, 15, 840–852. [Google Scholar] [CrossRef] [Green Version]
- Herzog, S.; Fink, M.A.; Weitmann, K.; Friedel, C.; Hadlich, S.; Langner, S.; Kindermann, K.; Holm, T.; Bohm, A.; Eskilsson, E.; et al. Pim1 kinase is upregulated in glioblastoma multiforme and mediates tumor cell survival. Neuro-Oncology 2015, 17, 223–242. [Google Scholar] [CrossRef]
- Weirauch, U.; Grunweller, A.; Cuellar, L.; Hartmann, R.K.; Aigner, A. U1 adaptors for the therapeutic knockdown of the oncogene pim-1 kinase in glioblastoma. Nucl. Acid Ther. 2013, 23, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.; Ruzevick, J.; Amin, A.G.; Lim, M. Potential role for STAT3 inhibitors in glioblastoma. Neurosurg. Clin. North Am. 2012, 23, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Ouedraogo, Z.G.; Biau, J.; Kemeny, J.L.; Morel, L.; Verrelle, P.; Chautard, E. Role of STAT3 in Genesis and Progression of Human Malignant Gliomas. Mol. Neurobiol. 2017, 54, 5780–5797. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Weirauch, U.; Ewe, A.; Uhmann, A.; Seifert, V.; Mittelbronn, M.; Harter, P.N.; Aigner, A.; Kogel, D. Therapeutic Targeting of Stat3 Using Lipopolyplex Nanoparticle-Formulated siRNA in a Syngeneic Orthotopic Mouse Glioma Model. Cancers 2019, 11, 333. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, A.; Eckerdt, F.; Bell, J.; Nakano, I.; Giles, F.J.; Cheng, S.Y.; Lulla, R.R.; Goldman, S.; Platanias, L.C. Targeting of glioblastoma cell lines and glioma stem cells by combined PIM kinase and PI3K-p110alpha inhibition. Oncotarget 2016, 7, 33192–33201. [Google Scholar] [CrossRef] [Green Version]
- Remy, J.; Linder, B.; Weirauch, U.; Konovalova, J.; Marschalek, R.; Aigner, A.; Kogel, D. Inhibition of PIM1 blocks the autophagic flux to sensitize glioblastoma cells to ABT-737-induced apoptosis. Biochimica Biophysica Acta Mol. Cell Res. 2019, 1866, 175–189. [Google Scholar] [CrossRef]
- Kunkel, P.; Ulbricht, U.; Bohlen, P.; Brockmann, M.A.; Fillbrandt, R.; Stavrou, D.; Westphal, M.; Lamszus, K. Inhibition of glioma angiogenesis and growth in vivo by systemic treatment with a monoclonal antibody against vascular endothelial growth factor receptor-2. Cancer Res. 2001, 61, 6624–6628. [Google Scholar]
- Hetschko, H.; Voss, V.; Horn, S.; Seifert, V.; Prehn, J.H.; Kögel, D. Pharmacological inhibition of Bcl-2 family members reactivates TRAIL-induced apoptosis in malignant glioma. J. Neuro-Oncol. 2008, 86, 265–272. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Hendruschk, S.; Wiedemuth, R.; Aigner, A.; Topfer, K.; Cartellieri, M.; Martin, D.; Kirsch, M.; Ikonomidou, C.; Schackert, G.; Temme, A. RNA interference targeting survivin exerts antitumoral effects in vitro and in established glioma xenografts in vivo. Neuro-Oncology 2011, 13, 1074–1089. [Google Scholar] [CrossRef] [Green Version]
- Heine, C.; Franke, H. Organotypic slice co-culture systems to study axon regeneration in the dopaminergic system ex vivo. Methods Mol. Biol. 2014, 1162, 97–111. [Google Scholar] [PubMed]
- McClelland, K.S.; Ng, E.T.; Bowles, J. Agarose/gelatin immobilisation of tissues or embryo segments for orientated paraffin embedding and sectioning. Differ. Res. Biol. Divers. 2016, 91, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.R.; Shi, Y.; Taylor, C.R. Antigen retrieval immunohistochemistry: Review and future prospects in research and diagnosis over two decades. J. Histochem. Cytochem. 2011, 59, 13–32. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sidorcenco, V.; Krahnen, L.; Schulz, M.; Remy, J.; Kögel, D.; Temme, A.; Krügel, U.; Franke, H.; Aigner, A. Glioblastoma Tissue Slice Tandem-Cultures for Quantitative Evaluation of Inhibitory Effects on Invasion and Growth. Cancers 2020, 12, 2707. https://doi.org/10.3390/cancers12092707
Sidorcenco V, Krahnen L, Schulz M, Remy J, Kögel D, Temme A, Krügel U, Franke H, Aigner A. Glioblastoma Tissue Slice Tandem-Cultures for Quantitative Evaluation of Inhibitory Effects on Invasion and Growth. Cancers. 2020; 12(9):2707. https://doi.org/10.3390/cancers12092707
Chicago/Turabian StyleSidorcenco, Vasile, Luisa Krahnen, Marion Schulz, Janina Remy, Donat Kögel, Achim Temme, Ute Krügel, Heike Franke, and Achim Aigner. 2020. "Glioblastoma Tissue Slice Tandem-Cultures for Quantitative Evaluation of Inhibitory Effects on Invasion and Growth" Cancers 12, no. 9: 2707. https://doi.org/10.3390/cancers12092707
APA StyleSidorcenco, V., Krahnen, L., Schulz, M., Remy, J., Kögel, D., Temme, A., Krügel, U., Franke, H., & Aigner, A. (2020). Glioblastoma Tissue Slice Tandem-Cultures for Quantitative Evaluation of Inhibitory Effects on Invasion and Growth. Cancers, 12(9), 2707. https://doi.org/10.3390/cancers12092707