Artificial Intelligence Applications to Improve the Treatment of Locally Advanced Non-Small Cell Lung Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
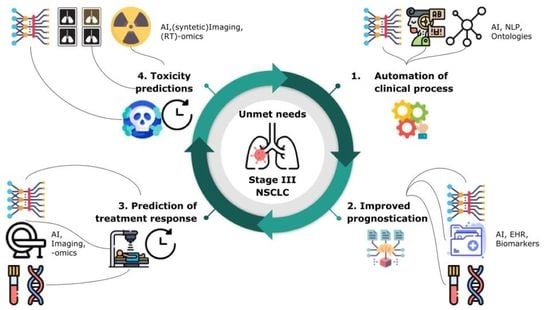
2. Unmet Clinical Needs in the Management of LA-NSCLC Patients: Role of Imaging, Adaptive RT, and Biomarkers
2.1. General Automation of Clinical Processes
- Contouring. As part of radiation treatment planning, contouring is a physician-directed image classification, whereby tumor targets (gross tumor volume (GTV) and clinical target volume (CTV)) are manually segmented as discrete and distinct from OAR or normal anatomy. Unfortunately, this is also a time-consuming process. AI has been shown to be capable of image classification in the clinical space [12,13,14,15], and rapid segmentation via AI represents a potential force-multiplier to enable individual radiation oncologists to evaluate and treat more patients per capita. This image-processing task has long been recognized as an important unmet clinical need, which has been a research target for numerous groups. Our work focused on the introduction in clinical practice of auto-contouring for RT OAR, comparing both atlas-based and deep learning algorithms [16], as well as on the automation of RT target volume delineations [17]. Recent results showed that deep learning algorithms can outperform expert technicians on lesion segmentation tasks [18].
- Automated review/classification of clinical records. Interestingly, as the digital domain for electronic medical records becomes the standard, the number and diverse types of clinical records accessible to AI for each patient is growing. However, the data in these resources are often unstructured (not discrete). As a result, there may be a role for natural language processing (NLP) in the review of clinical records, for example, to automatically extract comorbid illness, to identify pathologic diagnosis and/or biomarkers of relevance (epidermal growth factor receptor (EGFR), K-RAS, anaplastic lymphoma kinase (ALK), and programmed death ligand (PDL1)) from pathologic data, or to identify previous radiotherapy treatments that may identify risks for retreatment. We anticipate that the automated classification of free text medication records into a structured format will become increasingly important to monitor interactions between treatments and outcomes. For example, we have investigated the role of NLP in homogenizing radiological reports and in extracting standardized knowledge, such as the automated classification of tumor T stage from free text [19]. This work was extended to classify lesions as well as other characteristics from lung radiological reports. Despite promising results, the difficulties encountered during these studies underline the need for standardized nomenclature in medical records by the use of dedicated ontologies and semantic web techniques. This has been acknowledged by the European Society for Therapeutic Radiation Oncology (ESTRO) [20,21], the American Association of Physics in Medicine (AAPM) [22], and the American Society for Radiation Oncology (ASTRO) and resulted in the formation of working groups to establish these guidelines. We have also developed a deep learning system to automatically identify and extract tumor site and histology from free-text pathology reports. Our system predicts ICD-O-3 codes and preferred phrases with accuracies comparable to human experts [23]. In LA-NSCLC patients, it identifies lung subregions and tumor subtypes.
- Standardized data collection/ontological classification. There is also an interesting interaction between the automation described above and the subsequent usability of the data obtained. Automation not only leads to efficiency gains but also, generally, leads to more standardized/ontological data collection, which in turn may lead to better prognostication and prediction. As an example, a recent paper using AI-based automated heart segmentation led to a better prediction of dose-related cardiac toxicity in a pivotal trial on advanced lung cancer patients (RTOG 0617) compared to human heart segmentations, likely due to interobserver variation [24].
2.2. Improved Prognostication Regarding Expected Patient Outcomes in the Absence of Recurrent Disease
2.3. Characterization/Prediction of Malignant Disease Course: Disease Response to Treatments
2.4. Characterization/Prediction of Toxicity/Host Response to Treatment
2.5. Integration of all Predictive/Prognostic Metrics into Summary/Composite/Ensemble “Personalized” Prediction
3. AI in Medical Imaging: Dealing with Standard of Care Imaging and Confounding Factors. Are We AI Ready?
4. AI for Biomarker Discovery. from Medical Imaging to Biology
5. AI for ART Workflow Optimization
6. Deployment of Decision Support Systems
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Govindan, R.; Bogart, J.; Vokes, E.E. Locally Advanced Non-Small Cell Lung Cancer: The Past, Present, and Future. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2008, 3, 917–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, S.M.; Shaikh, T.; Hallman, M. Therapeutic Management Options for Stage III Non-Small Cell Lung Cancer. World J. Clin. Oncol. 2017, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, G.S.; Phillips, K.A. Precision Medicine: From Science to Value. Health Aff. Proj. Hope 2018, 37, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Huang, L.-L.; Chen, J.-H.; Wu, J.; Xu, Q. The Emerging Treatment Landscape of Targeted Therapy in Non-Small-Cell Lung Cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajappa, S.; Sharma, S.; Prasad, K. Unmet Clinical Need in the Management of Locally Advanced Unresectable Lung Cancer: Treatment Strategies to Improve Patient Outcomes. Adv. Ther. 2019, 36, 563–578. [Google Scholar] [CrossRef]
- Huber, R.M.; De Ruysscher, D.; Hoffmann, H.; Reu, S.; Tufman, A. Interdisciplinary Multimodality Management of Stage III Nonsmall Cell Lung Cancer. Eur. Respir. Rev. 2019, 28, 190024. [Google Scholar] [CrossRef]
- De Ruysscher, D.; Botterweck, A.; Dirx, M.; Pijls-Johannesma, M.; Wanders, R.; Hochstenbag, M.; Dingemans, A.-M.C.; Bootsma, G.; Geraedts, W.; Simons, J.; et al. Eligibility for Concurrent Chemotherapy and Radiotherapy of Locally Advanced Lung Cancer Patients: A Prospective, Population-Based Study. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2009, 20, 98–102. [Google Scholar] [CrossRef]
- Jalal, S.I.; Riggs, H.D.; Melnyk, A.; Richards, D.; Agarwala, A.; Neubauer, M.; Ansari, R.; Govindan, R.; Bruetman, D.; Fisher, W.; et al. Updated Survival and Outcomes for Older Adults with Inoperable Stage III Non-Small-Cell Lung Cancer Treated with Cisplatin, Etoposide, and Concurrent Chest Radiation with or without Consolidation Docetaxel: Analysis of a Phase III Trial from the Hoosier Oncology Group (HOG) and US Oncology. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, 1730–1738. [Google Scholar] [CrossRef]
- Bradley, J.D.; Hu, C.; Komaki, R.R.; Masters, G.A.; Blumenschein, G.R.; Schild, S.E.; Bogart, J.A.; Forster, K.M.; Magliocco, A.M.; Kavadi, V.S.; et al. Long-Term Results of NRG Oncology RTOG 0617: Standard- Versus High-Dose Chemoradiotherapy With or Without Cetuximab for Unresectable Stage III Non-Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 706–714. [Google Scholar] [CrossRef]
- Muñoz-Unceta, N.; Burgueño, I.; Jiménez, E.; Paz-Ares, L. Durvalumab in NSCLC: Latest Evidence and Clinical Potential. Ther. Adv. Med. Oncol. 2018, 10, 1758835918804151. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.D. Data Explosion: The Challenge of Multidetector-Row CT. Eur. J. Radiol. 2000, 36, 74–80. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Goyal, M.; Knackstedt, T.; Yan, S.; Hassanpour, S. Artificial Intelligence-Based Image Classification Methods for Diagnosis of Skin Cancer: Challenges and Opportunities. Comput. Biol. Med. 2020, 127, 104065. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Hauschild, A.; Weichenthal, M.; Maron, R.C.; Berking, C.; Haferkamp, S.; Klode, J.; Schadendorf, D.; et al. Superior Skin Cancer Classification by the Combination of Human and Artificial Intelligence. Eur. J. Cancer 2019, 120, 114–121. [Google Scholar] [CrossRef] [Green Version]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically Applicable Deep Learning for Diagnosis and Referral in Retinal Disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef]
- Lustberg, T.; van Soest, J.; Gooding, M.; Peressutti, D.; Aljabar, P.; van der Stoep, J.; van Elmpt, W.; Dekker, A. Clinical Evaluation of Atlas and Deep Learning Based Automatic Contouring for Lung Cancer. Radiother. Oncol. 2018, 126, 312–317. [Google Scholar] [CrossRef] [Green Version]
- Rios Velazquez, E.; Aerts, H.J.W.L.; Gu, Y.; Goldgof, D.B.; De Ruysscher, D.; Dekker, A.; Korn, R.; Gillies, R.J.; Lambin, P. A Semiautomatic CT-Based Ensemble Segmentation of Lung Tumors: Comparison with Oncologists’ Delineations and with the Surgical Specimen. Radiother. Oncol. 2012, 105, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, J.R.; Kamnitsas, K.; Singleton, K.W.; Whitmire, S.A.; Clark-Swanson, K.R.; Ranjbar, S.; Rickertsen, C.R.; Johnston, S.K.; Egan, K.M.; Rollison, D.E.; et al. Deep Neural Network to Locate and Segment Brain Tumors Outperformed the Expert Technicians Who Created the Training Data. J. Med. Imaging 2020, 7. [Google Scholar] [CrossRef]
- Nobel, J.M.; Puts, S.; Bakers, F.C.H.; Robben, S.G.F.; Dekker, A.L.A.J. Natural Language Processing in Dutch Free Text Radiology Reports: Challenges in a Small Language Area Staging Pulmonary Oncology. J. Digit. Imaging 2020, 33, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Kazmierska, J.; Hope, A.; Spezi, E.; Beddar, S.; Nailon, W.H.; Osong, B.; Ankolekar, A.; Choudhury, A.; Dekker, A.; Redalen, K.R.; et al. From Multisource Data to Clinical Decision Aids in Radiation Oncology: The Need for a Clinical Data Science Community. Radiother. Oncol. 2020, 153, 43–54. [Google Scholar] [CrossRef]
- Traverso, A.; van Soest, J.; Wee, L.; Dekker, A. The Radiation Oncology Ontology (ROO): Publishing Linked Data in Radiation Oncology Using Semantic Web and Ontology Techniques. Med. Phys. 2018, 45, e854–e862. [Google Scholar] [CrossRef]
- Mayo, C.S.; Moran, J.M.; Bosch, W.; Xiao, Y.; McNutt, T.; Popple, R.; Michalski, J.; Feng, M.; Marks, L.B.; Fuller, C.D.; et al. American Association of Physicists in Medicine Task Group 263: Standardizing Nomenclatures in Radiation Oncology. Int. J. Radiat. Oncol. 2018, 100, 1057–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, J.R.; Szepietowski, P.; Howard, R.; Reisman, P.; Jones, J.D.; Lewis, P.; Fridley, B.L.; Rollison, D.E. Cabernet: A Question-and-Answer System to Extract Data from Free-Text Pathology Reports (Preprint). J. Med. Internet Res. 2021. [Google Scholar] [CrossRef]
- Thor, M.; Apte, A.; Haq, R.; Iyer, A.; LoCastro, E.; Deasy, J.O. Using Auto-Segmentation to Reduce Contouring and Dose Inconsistency in Clinical Trials: The Simulated Impact on RTOG 0617. Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 1619–1626. [Google Scholar] [CrossRef]
- Korc-Grodzicki, B.; Holmes, H.M.; Shahrokni, A. Geriatric Assessment for Oncologists. Cancer Biol. Med. 2015, 12, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Calvani, R.; Marini, F.; Cesari, M.; Tosato, M.; Anker, S.D.; von Haehling, S.; Miller, R.R.; Bernabei, R.; Landi, F.; Marzetti, E.; et al. Biomarkers for Physical Frailty and Sarcopenia: State of the Science and Future Developments. J. Cachexia Sarcopenia Muscle 2015, 6, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Dunne, R.F.; Loh, K.P.; Williams, G.R.; Jatoi, A.; Mustian, K.M.; Mohile, S.G. Cachexia and Sarcopenia in Older Adults with Cancer: A Comprehensive Review. Cancers 2019, 11, 1861. [Google Scholar] [CrossRef] [Green Version]
- Grove, O.; Berglund, A.E.; Schabath, M.B.; Aerts, H.J.W.L.; Dekker, A.; Wang, H.; Velazquez, E.R.; Lambin, P.; Gu, Y.; Balagurunathan, Y.; et al. Quantitative Computed Tomographic Descriptors Associate Tumor Shape Complexity and Intratumor Heterogeneity with Prognosis in Lung Adenocarcinoma. PLoS ONE 2015, 10, e0118261. [Google Scholar] [CrossRef] [Green Version]
- Grossmann, P.; Stringfield, O.; El-Hachem, N.; Bui, M.M.; Rios Velazquez, E.; Parmar, C.; Leijenaar, R.T.; Haibe-Kains, B.; Lambin, P.; Gillies, R.J.; et al. Defining the Biological Basis of Radiomic Phenotypes in Lung Cancer. eLife 2017, 6, e23421. [Google Scholar] [CrossRef]
- Cherezov, D.; Goldgof, D.; Hall, L.; Gillies, R.; Schabath, M.; Müller, H.; Depeursinge, A. Revealing Tumor Habitats from Texture Heterogeneity Analysis for Classification of Lung Cancer Malignancy and Aggressiveness. Sci. Rep. 2019, 9, 4500. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, X.; Xu, K.; Qian, Z.; Wang, K.; Fan, X.; Li, S.; Wang, Y.; Jiang, T. MRI Features Can Predict EGFR Expression in Lower Grade Gliomas: A Voxel-Based Radiomic Analysis. Eur. Radiol. 2018, 28, 356–362. [Google Scholar] [CrossRef]
- Glick, D.; Lyen, S.; Kandel, S.; Shapera, S.; Le, L.W.; Lindsay, P.; Wong, O.; Bezjak, A.; Brade, A.; Cho, B.C.J.; et al. Impact of Pretreatment Interstitial Lung Disease on Radiation Pneumonitis and Survival in Patients Treated With Lung Stereotactic Body Radiation Therapy (SBRT). Clin. Lung Cancer 2018, 19, e219–e226. [Google Scholar] [CrossRef]
- Krafft, S.P.; Rao, A.; Stingo, F.; Briere, T.M.; Court, L.E.; Liao, Z.; Martel, M.K. The Utility of Quantitative CT Radiomics Features for Improved Prediction of Radiation Pneumonitis. Med. Phys. 2018, 45, 5317–5324. [Google Scholar] [CrossRef]
- Cunliffe, A.; Armato, S.G.; Castillo, R.; Pham, N.; Guerrero, T.; Al-Hallaq, H.A. Lung Texture in Serial Thoracic Computed Tomography Scans: Correlation of Radiomics-Based Features with Radiation Therapy Dose and Radiation Pneumonitis Development. Int. J. Radiat. Oncol. Biol. Phys. 2015, 91, 1048–1056. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Hosny, A.; Zeleznik, R.; Parmar, C.; Coroller, T.; Franco, I.; Mak, R.H.; Aerts, H.J.W.L. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3266–3275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhovannik, I.; Bussink, J.; Traverso, A.; Shi, Z.; Kalendralis, P.; Wee, L.; Dekker, A.; Fijten, R.; Monshouwer, R. Learning from Scanners: Bias Reduction and Feature Correction in Radiomics. Clin. Transl. Radiat. Oncol. 2019, 19, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [Green Version]
- van Timmeren, J.E.; Leijenaar, R.T.H.; van Elmpt, W.; Reymen, B.; Oberije, C.; Monshouwer, R.; Bussink, J.; Brink, C.; Hansen, O.; Lambin, P. Survival Prediction of Non-Small Cell Lung Cancer Patients Using Radiomics Analyses of Cone-Beam CT Images. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2017, 123, 363–369. [Google Scholar] [CrossRef] [Green Version]
- Ammari, S.; Pitre-Champagnat, S.; Dercle, L.; Chouzenoux, E.; Moalla, S.; Reuze, S.; Talbot, H.; Mokoyoko, T.; Hadchiti, J.; Diffetocq, S.; et al. Influence of Magnetic Field Strength on Magnetic Resonance Imaging Radiomics Features in Brain Imaging, an In Vitro and In Vivo Study. Front. Oncol. 2020, 10, 541663. [Google Scholar] [CrossRef]
- Ligero, M.; Jordi-Ollero, O.; Bernatowicz, K.; Garcia-Ruiz, A.; Delgado-Muñoz, E.; Leiva, D.; Mast, R.; Suarez, C.; Sala-Llonch, R.; Calvo, N.; et al. Minimizing Acquisition-Related Radiomics Variability by Image Resampling and Batch Effect Correction to Allow for Large-Scale Data Analysis. Eur. Radiol. 2020. [Google Scholar] [CrossRef]
- Lan, L.; You, L.; Zhang, Z.; Fan, Z.; Zhao, W.; Zeng, N.; Chen, Y.; Zhou, X. Generative Adversarial Networks and Its Applications in Biomedical Informatics. Front. Public Health 2020, 8, 164. [Google Scholar] [CrossRef]
- Maspero, M.; Houweling, A.C.; Savenije, M.H.F.; van Heijst, T.C.F.; Verhoeff, J.J.C.; Kotte, A.N.T.J.; van den Berg, C.A.T. A Single Neural Network for Cone-Beam Computed Tomography-Based Radiotherapy of Head-and-Neck, Lung and Breast Cancer. Phys. Imaging Radiat. Oncol. 2020, 14, 24–31. [Google Scholar] [CrossRef]
- Chalkidou, A.; O’Doherty, M.J.; Marsden, P.K. False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS ONE 2015, 10, e0124165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welch, M.L.; McIntosh, C.; Haibe-Kains, B.; Milosevic, M.F.; Wee, L.; Dekker, A.; Huang, S.H.; Purdie, T.G.; O’Sullivan, B.; Aerts, H.J.W.L.; et al. Vulnerabilities of Radiomic Signature Development: The Need for Safeguards. Radiother. Oncol. 2019, 130, 2–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traverso, A.; Kazmierski, M.; Zhovannik, I.; Welch, M.; Wee, L.; Jaffray, D.; Dekker, A.; Hope, A. Machine Learning Helps Identifying Volume-Confounding Effects in Radiomics. Phys. Med. 2020, 71, 24–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaupel, P.; Mayer, A. Hypoxia in Cancer: Significance and Impact on Clinical Outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef]
- Aran, V.; Omerovic, J. Current Approaches in NSCLC Targeting K-RAS and EGFR. Int. J. Mol. Sci. 2019, 20, 5701. [Google Scholar] [CrossRef] [Green Version]
- Paver, E.C.; Cooper, W.A.; Colebatch, A.J.; Ferguson, P.M.; Hill, S.K.; Lum, T.; Shin, J.-S.; O’Toole, S.; Anderson, L.; Scolyer, R.A.; et al. Programmed Death Ligand-1 (PD-L1) as a Predictive Marker for Immunotherapy in Solid Tumours: A Guide to Immunohistochemistry Implementation and Interpretation. Pathology 2021, 53, 141–156. [Google Scholar] [CrossRef]
- Palmer, J.D.; Zaorsky, N.G.; Witek, M.; Lu, B. Molecular Markers to Predict Clinical Outcome and Radiation Induced Toxicity in Lung Cancer. J. Thorac. Dis. 2014, 6, 387–398. [Google Scholar] [CrossRef]
- Sanduleanu, S.; Wiel, A.; Lieverse, R.I.; Marcus, D.; Ibrahim, A.; Primakov, S.; Wu, G.; Theys, J.; Yaromina, A.; Dubois, L.J.; et al. Hypoxia PET Imaging with [18F]-HX4—A Promising Next-Generation Tracer. Cancers 2020, 12, 1322. [Google Scholar] [CrossRef]
- Even, A.J.G.; Reymen, B.; La Fontaine, M.D.; Das, M.; Jochems, A.; Mottaghy, F.M.; Belderbos, J.S.A.; De Ruysscher, D.; Lambin, P.; van Elmpt, W. Predicting Tumor Hypoxia in Non-Small Cell Lung Cancer by Combining CT, FDG PET and Dynamic Contrast-Enhanced CT. Acta Oncol. 2017, 56, 1591–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcu, L.G.; Forster, J.C.; Bezak, E. The Potential Role of Radiomics and Radiogenomics in Patient Stratification by Tumor Hypoxia Status. J. Am. Coll. Radiol. 2019, 16, 1329–1337. [Google Scholar] [CrossRef]
- Sanduleanu, S.; Jochems, A.; Upadhaya, T.; Even, A.J.G.; Leijenaar, R.T.H.; Dankers, F.J.W.M.; Klaassen, R.; Woodruff, H.C.; Hatt, M.; Kaanders, H.J.A.M.; et al. Non-Invasive Imaging Prediction of Tumor Hypoxia: A Novel Developed and Externally Validated CT and FDG-PET-Based Radiomic Signatures. Radiother. Oncol. 2020, 153, 97–105. [Google Scholar] [CrossRef]
- Assessment of the Prognostic Value of Radiomic Features in 18 F-FMISO PET Imaging of Hypoxia in Postsurgery Brain Cancer Patients: Secondary Analysis of Imaging Data from a Single-Center Study and the Multicenter ACRIN 6684 Trial. Tomography 2020, 6, 14–22. [CrossRef] [PubMed]
- Crispin-Ortuzar, M.; Apte, A.; Grkovski, M.; Oh, J.H.; Lee, N.Y.; Schöder, H.; Humm, J.L.; Deasy, J.O. Predicting Hypoxia Status Using a Combination of Contrast-Enhanced Computed Tomography and [18F]-Fluorodeoxyglucose Positron Emission Tomography Radiomics Features. Radiother. Oncol. 2018, 127, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Veeraraghavan, H.; Friedman, C.F.; DeLair, D.F.; Ninčević, J.; Himoto, Y.; Bruni, S.G.; Cappello, G.; Petkovska, I.; Nougaret, S.; Nikolovski, I.; et al. Machine Learning-Based Prediction of Microsatellite Instability and High Tumor Mutation Burden from Contrast-Enhanced Computed Tomography in Endometrial Cancers. Sci. Rep. 2020, 10, 17769. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-L.; Tsuboi, M.; He, J.; John, T.; Grohe, C.; Majem, M.; Goldman, J.W.; Laktionov, K.; Kim, S.-W.; Kato, T.; et al. Osimertinib in Resected EGFR -Mutated Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef]
- Ninatti, G.; Kirienko, M.; Neri, E.; Sollini, M.; Chiti, A. Imaging-Based Prediction of Molecular Therapy Targets in NSCLC by Radiogenomics and AI Approaches: A Systematic Review. Diagnostics 2020, 10, 359. [Google Scholar] [CrossRef]
- Rossi, G.; Barabino, E.; Fedeli, A.; Ficarra, G.; Coco, S.; Russo, A.; Adamo, V.; Buemi, F.; Zullo, L.; Dono, M.; et al. Radiomic Detection of EGFR Mutations in NSCLC. Cancer Res. 2021, 81, 724–731. [Google Scholar] [CrossRef]
- Zhang, M.; Bao, Y.; Rui, W.; Shangguan, C.; Liu, J.; Xu, J.; Lin, X.; Zhang, M.; Huang, X.; Zhou, Y.; et al. Performance of 18F-FDG PET/CT Radiomics for Predicting EGFR Mutation Status in Patients with Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 568857. [Google Scholar] [CrossRef]
- Nair, J.K.R.; Saeed, U.A.; McDougall, C.C.; Sabri, A.; Kovacina, B.; Raidu, B.V.S.; Khokhar, R.A.; Probst, S.; Hirsh, V.; Chankowsky, J.; et al. Radiogenomic Models Using Machine Learning Techniques to Predict EGFR Mutations in Non-Small Cell Lung Cancer. Can. Assoc. Radiol. J. 2021, 72, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, L.; Ng, N.N.; Zhao, M.; Shi, J.; Wu, N.; Li, W.; Liu, Z.; Yeom, K.W.; Tian, J. Development and Validation of a Machine Learning Model to Explore Tyrosine Kinase Inhibitor Response in Patients with Stage IV EGFR Variant–Positive Non–Small Cell Lung Cancer. JAMA Netw. Open 2020, 3, e2030442. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, R.; Wang, S.; Fang, M.; Zhu, Y.; Hu, Z.; Dong, D.; Shi, J.; Tian, J. CT-Based Radiomic Signature as a Prognostic Factor in Stage IV ALK-Positive Non-Small-Cell Lung Cancer Treated with TKI Crizotinib: A Proof-of-Concept Study. Front. Oncol. 2020, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Yokoi, T.; Chiappori, A.; Lee, K.H.; de Wit, M.; et al. Durvalumab after Chemoradiotherapy in Stage III Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 1919–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, B.; Dong, D.; She, Y.; Zhou, C.; Fang, M.; Zhu, Y.; Zhang, H.; Huang, Z.; Jiang, T.; Tian, J.; et al. Predicting Response to Immunotherapy in Advanced Non-Small-Cell Lung Cancer Using Tumor Mutational Burden Radiomic Biomarker. J. Immunother. Cancer 2020, 8, e000550. [Google Scholar] [CrossRef]
- Jiang, M.; Sun, D.; Guo, Y.; Guo, Y.; Xiao, J.; Wang, L.; Yao, X. Assessing PD-L1 Expression Level by Radiomic Features From PET/CT in Nonsmall Cell Lung Cancer Patients: An Initial Result. Acad. Radiol. 2020, 27, 171–179. [Google Scholar] [CrossRef]
- Sun, Z.; Hu, S.; Ge, Y.; Wang, J.; Duan, S.; Song, J.; Hu, C.; Li, Y. Radiomics Study for Predicting the Expression of PD-L1 in Non-Small Cell Lung Cancer Based on CT Images and Clinicopathologic Features. J. X-ray Sci. Technol. 2020, 28, 449–459. [Google Scholar] [CrossRef]
- Tunali, I.; Gray, J.E.; Qi, J.; Abdalah, M.; Jeong, D.K.; Guvenis, A.; Gillies, R.J.; Schabath, M.B. Novel Clinical and Radiomic Predictors of Rapid Disease Progression Phenotypes among Lung Cancer Patients Treated with Immunotherapy: An Early Report. Lung Cancer 2019, 129, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Tunali, I.; Gray, J.E.; Qi, J.; Schabath, M.B.; Gillies, R.J. Radiomics of 18F-FDG PET/CT Images Predicts Clinical Benefit of Advanced NSCLC Patients to Checkpoint Blockade Immunotherapy. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1168–1182. [Google Scholar] [CrossRef]
- Vaidya, P.; Bera, K.; Patil, P.D.; Gupta, A.; Jain, P.; Alilou, M.; Khorrami, M.; Velcheti, V.; Madabhushi, A. Novel, Non-Invasive Imaging Approach to Identify Patients with Advanced Non-Small Cell Lung Cancer at Risk of Hyperprogressive Disease with Immune Checkpoint Blockade. J. Immunother. Cancer 2020, 8, e001343. [Google Scholar] [CrossRef]
- El Naqa, I.; Haider, M.A.; Giger, M.L.; Ten Haken, R.K. Artificial Intelligence: Reshaping the Practice of Radiological Sciences in the 21st Century. Br. J. Radiol. 2020, 93, 20190855. [Google Scholar] [CrossRef] [PubMed]
- Palma, G.; Monti, S.; Conson, M.; Pacelli, R.; Cella, L. Normal Tissue Complication Probability (NTCP) Models for Modern Radiation Therapy. Semin. Oncol. 2019, 46, 210–218. [Google Scholar] [CrossRef]
- Tseng, H.-H.; Luo, Y.; Cui, S.; Chien, J.-T.; Ten Haken, R.K.; Naqa, I.E. Deep Reinforcement Learning for Automated Radiation Adaptation in Lung Cancer. Med. Phys. 2017, 44, 6690–6705. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From Local Explanations to Global Understanding with Explainable AI for Trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Goodman, B.; Flaxman, S. European Union Regulations on Algorithmic Decision-Making and a “Right to Explanation”. AI Mag. 2017, 38, 50–57. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hope, A.; Verduin, M.; Dilling, T.J.; Choudhury, A.; Fijten, R.; Wee, L.; Aerts, H.J.; El Naqa, I.; Mitchell, R.; Vooijs, M.; et al. Artificial Intelligence Applications to Improve the Treatment of Locally Advanced Non-Small Cell Lung Cancers. Cancers 2021, 13, 2382. https://doi.org/10.3390/cancers13102382
Hope A, Verduin M, Dilling TJ, Choudhury A, Fijten R, Wee L, Aerts HJ, El Naqa I, Mitchell R, Vooijs M, et al. Artificial Intelligence Applications to Improve the Treatment of Locally Advanced Non-Small Cell Lung Cancers. Cancers. 2021; 13(10):2382. https://doi.org/10.3390/cancers13102382
Chicago/Turabian StyleHope, Andrew, Maikel Verduin, Thomas J Dilling, Ananya Choudhury, Rianne Fijten, Leonard Wee, Hugo JWL Aerts, Issam El Naqa, Ross Mitchell, Marc Vooijs, and et al. 2021. "Artificial Intelligence Applications to Improve the Treatment of Locally Advanced Non-Small Cell Lung Cancers" Cancers 13, no. 10: 2382. https://doi.org/10.3390/cancers13102382
APA StyleHope, A., Verduin, M., Dilling, T. J., Choudhury, A., Fijten, R., Wee, L., Aerts, H. J., El Naqa, I., Mitchell, R., Vooijs, M., Dekker, A., de Ruysscher, D., & Traverso, A. (2021). Artificial Intelligence Applications to Improve the Treatment of Locally Advanced Non-Small Cell Lung Cancers. Cancers, 13(10), 2382. https://doi.org/10.3390/cancers13102382