Computational Treatment Simulations to Assess the Need for Personalized Tamoxifen Dosing in Breast Cancer Patients of Different Biogeographical Groups
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Virtual Patients of Different Biogeographical Groups
2.2. Joint Parent–Metabolite Pharmacokinetic Model of Tamoxifen and Endoxifen and Simulations
3. Results
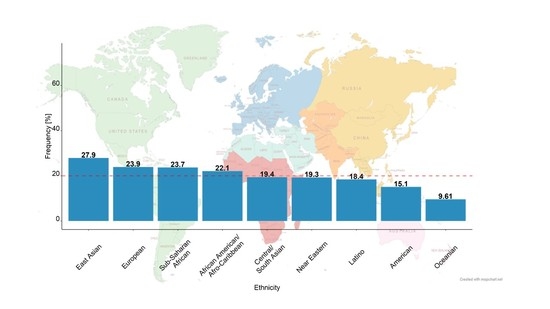
3.1. Virtual Patients of Different Biogeographical Groups
- American: e.g., Alaska Natives, American Native Indians, Argentinians, Canadians (Inuit and Native Indians), Chileans (Mapuches), Costa Ricans (Amerindians), Ecuadorians, Mexicans (Amerindians and Mexican Natives), Native Americans from South, North, and Central America as well as Panamanian, Paraguayan, and Venezuelan populations
- African American/Afro-Caribbean: e.g., African American, Antillean, Costa Rican, Cuban, Trinidadian
- Central/South Asian: e.g., Indian, Pakistani, Tamil, and Trinidadian populations
- East Asian: e.g., Chinese, Filipinos, Japanese, Karen, Korean, Malay, Mongolian, Russian (Russian Far East), Thai, Tibetan, Uyghur, and Vietnamese populations
- European: e.g., Albanian, Austrian, Belgian, Brazilian (of European descent), Caucasian, Croatian, Cuban (of European descent), Czech, Danish, Dutch, Faroese, Finnish, French, German, Greek, Hungarian, Italian, Macedonian, North American, Norwegian, Polish, Portuguese, Roma, Russian (Voronezh Region + St. Petersburg), Sardinian, Spanish, Swedish, and Swiss populations
- Latino: e.g., including Admixed Latin American, Hispanic American, Brazilian, Chilean, Columbian, Costa Rican, Cuban, Ecuadorian (Mestizos), Mexican, Nicaraguan, Puerto Rican, and Venezuelan populations
- Near Eastern: e.g., Ashkenazi Jews, Bedouin, Emirati, Iranian, Iraqi, Middle-Eastern, Saudi Arabian, Syrian, and Turkish populations
- Oceanian: e.g., Australian Aborigines, Maori, Papua New Guinean, Hawaiian, Melanesian, and Polynesian populations
- Sub-Saharan African: e.g., Brazilian (with African descent), Ethiopian, Ghanaian, South African, Tanzanian, Xhosa, Venda, Zimbabwean, and Kenyan populations.
3.2. Simulated Endoxifen Steady-State Concentrations at Tamoxifen Standard Dosing for Each Biogeographic Group
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, O.; Abe, R.; Enomoto, K.; Kikuchi, K.; Koyama, H.; Masuda, H.; Nomura, Y.; Ohashi, Y.; Sakai, K.; Sugimachi, K.; et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.; Zackrisson, S.; Senkus, E. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burstein, H.J.; Lacchetti, C.; Anderson, H.; Buchholz, T.A.; Davidson, N.E.; Gelmon, K.A.; Giordano, S.H.; Hudis, C.A.; Solky, A.J.; Stearns, V.; et al. Adjuvant Endocrine Therapy for Women With Hormone Receptor–Positive Breast Cancer: ASCO Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2019, 37, 423–438. [Google Scholar] [CrossRef]
- Johnson, M.D.; Zuo, H.; Lee, K.-H.; Trebley, J.P.; Rae, J.M.; Weatherman, R.V.; Desta, Z.; Flockhart, D.A.; Skaar, T.C. Pharmacological Characterization of 4-hydroxy-N-desmethyl Tamoxifen, a Novel Active Metabolite of Tamoxifen. Breast Cancer Res. Treat. 2004, 85, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Mürdter, T.E.; Schroth, W.; Bacchus-Gerybadze, L.; Winter, S.; Heinkele, G.; Simon, W.; Fasching, P.A.; Fehm, T.; Eichelbaum, M.; Schwab, M.; et al. Activity Levels of Tamoxifen Metabolites at the Estrogen Receptor and the Impact of Genetic Polymorphisms of Phase I and II Enzymes on Their Concentration Levels in Plasma. Clin. Pharmacol. Ther. 2011, 89, 708–717. [Google Scholar] [CrossRef]
- Sanchez-Spitman, A.; Swen, J.; Dezentje, V.; Moes, D.; Gelderblom, H.; Guchelaar, H. Clinical pharmacokinetics and pharmacogenetics of tamoxifen and endoxifen. Expert Rev. Clin. Pharmacol. 2019, 12, 523–536. [Google Scholar] [CrossRef]
- Saladores, P.H.; Murdter, T.E.; Eccles, D.; Chowbay, B.; Zgheib, N.K.; Winter, S.; Ganchev, B.; Eccles, B.K.; Gerty, S.; Tfayli, A.; et al. Tamoxifen metabolism predicts drug concentrations and outcome in premenopausal patients with early breast cancer. Pharmacogenom. J. 2015, 15, 84–94. [Google Scholar] [CrossRef] [Green Version]
- Madlensky, L.; Natarajan, L.; Tchu, S.; Pu, M.; Mortimer, J.; Flatt, S.W.; Nikoloff, D.M.; Hillman, G.; Fontecha, M.R.; Lawrence, H.J.; et al. Tamoxifen Metabolite Concentrations, CYP2D6 Genotype, and Breast Cancer Outcomes. Clin. Pharmacol. Ther. 2011, 89, 718–725. [Google Scholar] [CrossRef]
- Desta, Z.; Ward, B.A.; Soukhova, N.V.; Flockhart, D.A. Comprehensive Evaluation of Tamoxifen Sequential Biotransformation by the Human Cytochrome P450 System in Vitro: Prominent Roles for CYP3A and CYP2D6. J. Pharmacol. Exp. Ther. 2004, 310, 1062–1075. [Google Scholar] [CrossRef]
- Klopp-Schulze, L.; Mueller-Schoell, A.; Neven, P.; Koolen, S.L.W.; Mathijssen, R.H.J.; Joerger, M.; Kloft, C. Integrated Data Analysis of Six Clinical Studies Points Toward Model-Informed Precision Dosing of Tamoxifen. Front. Pharmacol. 2020, 11, 283. [Google Scholar] [CrossRef]
- Mueller-Schoell, A.; Klopp-Schulze, L.; Schroth, W.; Mürdter, T.; Michelet, R.; Brauch, H.; Huisinga, W.; Joerger, M.; Neven, P.; Koolen, S.L.; et al. Obesity Alters Endoxifen Plasma Levels in Young Breast Cancer Patients: A Pharmacometric Simulation Approach. Clin. Pharmacol. Ther. 2020, 108, 661–670. [Google Scholar] [CrossRef]
- Wu, A.H.; Pike, M.C.; Williams, L.D.; Spicer, D.; Tseng, C.-C.; Churchwell, M.I.; Doerge, D.R. Tamoxifen, Soy, and Lifestyle Factors in Asian American Women With Breast Cancer. J. Clin. Oncol. 2007, 25, 3024–3030. [Google Scholar] [CrossRef]
- Peyrade, F.; Frenay, M.; Etienne, M.-C.; Ruch, F.; Guillemare, C.; Francois, E.; Namer, M.; Ferrero, J.-M.; Milano, G. Age-related difference in tamoxifen disposition. Clin. Pharmacol. Ther. 1996, 59, 401–410. [Google Scholar] [CrossRef]
- Lien, E.A.; Søiland, H.; Lundgren, S.; Aas, T.; Steen, V.M.; Mellgren, G.; Gjerde, J. Serum concentrations of tamoxifen and its metabolites increase with age during steady-state treatment. Breast Cancer Res. Treat. 2013, 141, 243–248. [Google Scholar] [CrossRef] [Green Version]
- Ximenez, J.P.B.; De Andrade, J.M.; Marques, M.P.; Coelho, E.B.; Suarez-Kurtz, G.; Lanchote, V.L. Hormonal status affects plasma exposure of tamoxifen and its main metabolites in tamoxifen-treated breast cancer patients. BMC Pharmacol. Toxicol. 2019, 20, 1–10. [Google Scholar] [CrossRef]
- Nardin, J.M.; Schroth, W.; Almeida, T.A.; Mürdter, T.; Picolotto, S.; Vendramini, E.C.L.; Hoppe, R.; Kogin, J.P.; Miqueleto, D.; De Moraes, S.D.R.; et al. The Influences of Adherence to Tamoxifen and CYP2D6 Pharmacogenetics on Plasma Concentrations of the Active Metabolite (Z)-Endoxifen in Breast Cancer. Clin. Transl. Sci. 2019, 13, 284–292. [Google Scholar] [CrossRef] [Green Version]
- Pistilli, B.; Paci, A.; Ferreira, A.R.; Di Meglio, A.; Poinsignon, V.; Bardet, A.; Menvielle, G.; Dumas, A.; Pinto, S.; Dauchy, S.; et al. Serum Detection of Nonadherence to Adjuvant Tamoxifen and Breast Cancer Recurrence Risk. J. Clin. Oncol. 2020, 38, 2762–2772. [Google Scholar] [CrossRef]
- He, W.; Grassmann, F.; Eriksson, M.; Eliasson, E.; Margolin, S.; Thorén, L.; Hall, P.; Czene, K. CYP2D6 Genotype Predicts Tamoxifen Discontinuation and Prognosis in Patients With Breast Cancer. J. Clin. Oncol. 2020, 38, 548–557. [Google Scholar] [CrossRef]
- Nofziger, C.; Turner, A.J.; Sangkuhl, K.; Whirl-Carrillo, M.; Agúndez, J.A.G.; Black, J.L.; Dunnenberger, H.M.; Ruano, G.; Kennedy, M.A.; Phillips, M.S.; et al. PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 2020, 107, 154–170. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Wu, H.-W.; Zhang, Y.-H. Effects of CYP2D6*10 polymorphism on tamoxifen pharmacokinetics in patients with breast cancer in Asia: A meta-analysis. Cancer Chemother. Pharmacol. 2018, 83, 71–79. [Google Scholar] [CrossRef]
- Lu, J.; Li, H.; Guo, P.; Shen, R.; Luo, Y.; Ge, Q.; Shi, W.; Li, Y.; Zhu, W. The effect of CYP2D6 *10 polymorphism on adjuvant tamoxifen in Asian breast cancer patients: A meta-analysis. OncoTargets Ther. 2017, 10, 5429–5437. [Google Scholar] [CrossRef] [Green Version]
- Chin, F.W.; Chan, S.C.; Rahman, S.A.; Akmal, S.N.; Rosli, R. CYP2D6 Genetic Polymorphisms and Phenotypes in Different Ethnicities of Malaysian Breast Cancer Patients. Breast J. 2016, 22, 54–62. [Google Scholar] [CrossRef]
- Charoenchokthavee, W.; Panomvana, D.; Sriuranpong, V.; Areepium, N. Prevalence of CYP2D6*2, CYP2D6*4, CYP2D6*10, and CYP3A5*3 in Thai breast cancer patients undergoing tamoxifen treatment. Breast Cancer Targets Ther. 2016, 8, 149–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panigoro, S.S.; Purwanto, D.J.; Hidayat, A.; Louisa, M.; Andalusia, R.; Setiabudy, R. Association of CYP2D6*10 (c. 100 C>T) Genotype with Z-END Concentration in Patients with Breast Cancer Receiving Tamoxifen Therapy in Indonesian Population. Endoc. Metab. Immune Disord. Drug Targets 2019, 19, 1198–1206. [Google Scholar] [CrossRef]
- Li, H.; Feng, L.; Xu, Y.; Yao, L.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Lin, B.; Li, J.; et al. The association of CYP2D6 *10 polymorphism with breast cancer risk and clinico-pathologic characteristics in Chinese women. Acta Oncol. 2006, 45, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Nazir, N.; Waheed, A.; Farhat, K.; Ismail, M.; Mansoor, Q. Frequency of CYP2D6*10 genotypes in Pakistani breast cancer patients taking adjuvant tamoxifen. J. Pak. Med. Assoc. 2016, 66, 1554–1558. [Google Scholar] [PubMed]
- Huddart, R.; Fohner, A.E.; Whirl-Carrillo, M.; Wojcik, G.L.; Gignoux, C.R.; Popejoy, A.B.; Bustamante, C.D.; Altman, R.B.; Klein, T.E. Standardized Biogeographic Grouping System for Annotating Populations in Pharmacogenetic Research. Clin. Pharmacol. Ther. 2019, 105, 1256–1262. [Google Scholar] [CrossRef]
- CPIC CYP2D6 Frequency Table. Available online: https://api.pharmgkb.org/v1/download/file/attachment/CYP2D6_frequency_table.xlsx (accessed on 19 January 2021).
- Klopp-Schulze, L.; Joerger, M.; Wicha, S.G.; Ter Heine, R.; Csajka, C.; Parra-Guillen, Z.P.; Kloft, C. Exploiting Pharmacokinetic Models of Tamoxifen and Endoxifen to Identify Factors Causing Subtherapeutic Concentrations in Breast Cancer Patients. Clin. Pharmacokinet. 2017, 57, 229–242. [Google Scholar] [CrossRef]
- Mueller-Schoell, A.; Klopp-Schulze, L.; Michelet, R.; van Dyk, M.; Mürdter, T.; Schwab, M.; Joerger, M.; Huisinga, W.; Mikus, G.; Kloft, C. Simulation-Based Assessment of the Impact of Non-Adherence on Endoxifen Target Attainment in Different Tamoxifen Dosing Strategies. Pharmaceuticals 2021, 14, 115. [Google Scholar] [CrossRef]
- PharmGKB Biogeographical Groups. Available online: https://www.pharmgkb.org/page/biogeographicalGroups (accessed on 19 January 2021).
- Gaedigk, A.; Simon, S.D.; Pearce, R.E.; Bradford, L.D.; Kennedy, M.J.; Leeder, J.S. The CYP2D6 Activity Score: Translating Genotype Information into a Qualitative Measure of Phenotype. Clin. Pharmacol. Ther. 2008, 83, 234–242. [Google Scholar] [CrossRef]
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.-J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777. [Google Scholar] [CrossRef] [Green Version]
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP 2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2019, 13, 116–124. [Google Scholar] [CrossRef] [Green Version]
- Keizer, R.J.; Karlsson, M.O.; Hooker, A. Modeling and Simulation Workbench for NONMEM: Tutorial on Pirana, PsN, and Xpose. CPT Pharmacometr. Syst. Pharmacol. 2013, 2, 50–59. [Google Scholar] [CrossRef]
- Wang, T.; Zhou, Y.; Cao, G. Pharmacogenetics of tamoxifen therapy in Asian populations: From genetic polymorphism to clinical outcomes. Eur. J. Clin. Pharmacol. 2021, 1–17. [Google Scholar] [CrossRef]
- Hertz, D.L.; Deal, A.; Ibrahim, J.G.; Walko, C.M.; Weck, K.E.; Anderson, S.; Magrinat, G.; Olajide, O.; Moore, S.; Raab, R.; et al. Tamoxifen Dose Escalation in Patients with Diminished CYP2D6 Activity Normalizes Endoxifen Concentrations Without Increasing Toxicity. Oncologist 2016, 21, 795–803. [Google Scholar] [CrossRef] [Green Version]
- Welzen, M.E.B.; Dezentjé, V.O.; Van Schaik, R.H.N.; Colbers, A.P.H.; Guchelaar, H.-J.; Van Erp, N.P.; Hartigh, J.D.; Burger, D.M.; Van Laarhoven, H.W.M. The Effect of Tamoxifen Dose Increment in Patients With Impaired CYP2D6 Activity. Ther. Drug Monit. 2015, 37, 501–507. [Google Scholar] [CrossRef]
- Fox, P.; Balleine, R.L.; Lee, C.; Gao, B.; Balakrishnar, B.; Menzies, A.M.; Yeap, S.H.; Ali, S.S.; Gebski, V.; Provan, P.J.; et al. Dose Escalation of Tamoxifen in Patients with Low Endoxifen Level: Evidence for Therapeutic Drug Monitoring—The TADE Study. Clin. Cancer Res. 2016, 22, 3164–3171. [Google Scholar] [CrossRef] [Green Version]
- Khalaj, Z.; Baratieh, Z.; Nikpour, P.; Schwab, M.; Schaeffeler, E.; Mokarian, F.; Khanahmad, H.; Salehi, R.; Mürdter, T.E.; Salehi, M. Clinical Trial: CYP2D6 Related Dose Escalation of Tamoxifen in Breast Cancer Patients With Iranian Ethnic Background Resulted in Increased Concentrations of Tamoxifen and Its Metabolites. Front. Pharmacol. 2019, 10, 530. [Google Scholar] [CrossRef]
- Shah, R.R.; Smith, R.L. Addressing phenoconversion: The Achilles’ heel of personalized medicine. Br. J. Clin. Pharmacol. 2015, 79, 222–240. [Google Scholar] [CrossRef] [Green Version]
- Gusella, M.; Pasini, F.; Corso, B.; Bertolaso, L.; De Rosa, G.; Falci, C.; Modena, Y.; Barile, C.; Da Corte, Z.D.; Fraccon, A.; et al. Predicting steady-state endoxifen plasma concentrations in breast cancer patients by CYP2D6 genotyping or phenotyping. Which approach is more reliable? Pharmacol. Res. Perspect. 2020, 8, e00646. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.I.; Low, S.K.; Maldonado, R.; Fox, P.; Balakrishnar, B.; Coulter, S.; de Bruijn, P.; Koolen, S.L.; Gao, B.; Lynch, J.; et al. Simplified phenotyping of CYP2D6 for tamoxifen treatment using the N-desmethyl-tamoxifen/ endoxifen ratio. Breast 2020, 54, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Binkhorst, L.; Mathijssen, R.H.; Jager, A.; van Gelder, T. Individualization of tamoxifen therapy: Much more than just CYP2D6 genotyping. Cancer Treat. Rev. 2015, 41, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Kluwe, F.; Michelet, R.; Mueller-Schoell, A.; Maier, C.; Klopp-Schulze, L.; Van Dyk, M.; Mikus, G.; Huisinga, W.; Kloft, C. Perspectives on Model-Informed Precision Dosing in the Digital Health Era: Challenges, Opportunities, and Recommendations. Clin. Pharmacol. Ther. 2021, 109, 29–36. [Google Scholar] [CrossRef]
- Sanchez-Spitman, A.; Dezentjé, V.; Swen, J.; Moes, D.J.A.R.; Böhringer, S.; Batman, E.; van Druten, E.; Smorenburg, C.; van Bochove, A.; Zeillemaker, A.; et al. Tamoxifen Pharmacogenetics and Metabolism: Results From the Prospective CYPTAM Study. J. Clin. Oncol. 2019, 37, 636–646. [Google Scholar] [CrossRef]
- Regan, M.M.; Leyland-Jones, B.; Bouzyk, M.; Pagani, O.; Tang, W.; Kammler, R.; Dell’Orto, P.; Biasi, M.O.; Thürlimann, B.; Lyng, M.B.; et al. CYP2D6 Genotype and Tamoxifen Response in Postmenopausal Women with Endocrine-Responsive Breast Cancer: The Breast International Group 1-98 Trial. J. Natl. Cancer Inst. 2012, 104, 441–451. [Google Scholar] [CrossRef]
- Rae, J.M.; Drury, S.; Hayes, D.F.; Stearns, V.; Thibert, J.N.; Haynes, B.P.; Salter, J.; Sestak, I.; Cuzick, J.; Dowsett, M. CYP2D6 and UGT2B7 Genotype and Risk of Recurrence in Tamoxifen-Treated Breast Cancer Patients. J. Natl. Cancer Inst. 2012, 104, 452–460. [Google Scholar] [CrossRef]
- Ratain, M.J.; Nakamura, Y.; Cox, N.J. CYP2D6 Genotype and Tamoxifen Activity: Understanding Interstudy Variability in Methodological Quality. Clin. Pharmacol. Ther. 2013, 94, 185–187. [Google Scholar] [CrossRef] [Green Version]
- Braal, C.L.; Beijnen, J.H.; Koolen, S.L.; Hoop, E.O.-D.; Steeghs, N.; Jager, A.; Huitema, A.D.; Mathijssen, R.H. Relevance of Endoxifen Concentrations: Absence of Evidence Is Not Evidence of Absence. J. Clin. Oncol. 2019, 37, 1980–1981. [Google Scholar] [CrossRef]
- de Vries Schultink, A.H.M.; Dorlo, T.P.C.; Madlensky, L.; Pierce, J.P.; Beijnen, J.H.; Huitema, A.D.R. Prospective Evaluation of Therapeutic Drug Monitoring of Endoxifen: Feasibility of Observational and Randomized Trials. Available online: http://page-meeting.org/?abstract=9150 (accessed on 1 December 2019).
- Helland, T.; Henne, N.; Bifulco, E.; Naume, B.; Borgen, E.; Kristensen, V.N.; Kvaløy, J.T.; Lash, T.L.; Alnæs, G.I.G.; Van Schaik, R.H.; et al. Serum concentrations of active tamoxifen metabolites predict long-term survival in adjuvantly treated breast cancer patients. Breast Cancer Res. 2017, 19, 1–13. [Google Scholar] [CrossRef]
- De Censi, A.; Johansson, H.; Helland, T.; Puntoni, M.; Macis, D.; Aristarco, V.; Caviglia, S.; Webber, T.B.; Briata, I.M.; D’Amico, M.; et al. Association of CYP2D6 genotype and tamoxifen metabolites with breast cancer recurrence in a low-dose trial. NPJ Breast Cancer 2021, 7, 1–5. [Google Scholar] [CrossRef]
- Vay, M.; Meyer, M.J.; Blank, A.; Skopp, G.; Rose, P.; Tzvetkov, M.V.; Mikus, G. Oral Yohimbine as a New Probe Drug to Predict CYP2D6 Activity: Results of a Fixed-Sequence Phase I Trial. Clin. Pharmacokinet. 2020, 59, 927–939. [Google Scholar] [CrossRef] [Green Version]



| Virtual Population | AS 0 | AS 0.5 | AS 1 | AS 1.5 | AS 2 1 | AS > 2 |
|---|---|---|---|---|---|---|
| American | 2.18% | 1.46% | 22.1% | 7.24% | 61.4% | 5.61% |
| African American/Afro-Caribbean | 2.33% | 10.5% | 25.8% | 32.1% | 24.7% | 4.67% |
| Central/South Asian | 2.34% | 7.36% | 22.2% | 25.8% | 39.9% | 2.48% |
| East Asian | 0.865% | 27.9% | 11.2% | 36.6% | 22.0% | 1.38% |
| European | 6.47% | 7.52% | 31.4% | 17.0% | 34.5% | 3.13% |
| Latino | 3.12% | 4.77% | 24.3% | 17.0% | 46.2% | 4.58% |
| Near Eastern | 2.20% | 8.34% | 21.5% | 26.7% | 30.9% | 10.4% |
| Oceanian | 0.383% | 0.482% | 9.61% | 5.22% | 63.7% | 20.6% |
| Sub-Saharan African | 1.53% | 12.3% | 31.2% | 31.7% | 18.6% | 4.70% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mueller-Schoell, A.; Michelet, R.; Klopp-Schulze, L.; van Dyk, M.; Mürdter, T.E.; Schwab, M.; Joerger, M.; Huisinga, W.; Mikus, G.; Kloft, C. Computational Treatment Simulations to Assess the Need for Personalized Tamoxifen Dosing in Breast Cancer Patients of Different Biogeographical Groups. Cancers 2021, 13, 2432. https://doi.org/10.3390/cancers13102432
Mueller-Schoell A, Michelet R, Klopp-Schulze L, van Dyk M, Mürdter TE, Schwab M, Joerger M, Huisinga W, Mikus G, Kloft C. Computational Treatment Simulations to Assess the Need for Personalized Tamoxifen Dosing in Breast Cancer Patients of Different Biogeographical Groups. Cancers. 2021; 13(10):2432. https://doi.org/10.3390/cancers13102432
Chicago/Turabian StyleMueller-Schoell, Anna, Robin Michelet, Lena Klopp-Schulze, Madelé van Dyk, Thomas E. Mürdter, Matthias Schwab, Markus Joerger, Wilhelm Huisinga, Gerd Mikus, and Charlotte Kloft. 2021. "Computational Treatment Simulations to Assess the Need for Personalized Tamoxifen Dosing in Breast Cancer Patients of Different Biogeographical Groups" Cancers 13, no. 10: 2432. https://doi.org/10.3390/cancers13102432
APA StyleMueller-Schoell, A., Michelet, R., Klopp-Schulze, L., van Dyk, M., Mürdter, T. E., Schwab, M., Joerger, M., Huisinga, W., Mikus, G., & Kloft, C. (2021). Computational Treatment Simulations to Assess the Need for Personalized Tamoxifen Dosing in Breast Cancer Patients of Different Biogeographical Groups. Cancers, 13(10), 2432. https://doi.org/10.3390/cancers13102432