Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma
Abstract
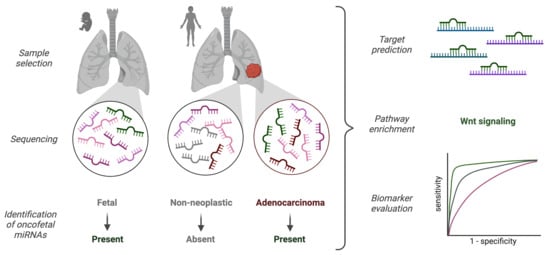
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cohort Composition
2.2. Quantification of miRNA Expression
2.3. Assessment of Sample-Type Specificity
2.4. Characterization of Differential and Oncofetal Expression
2.5. Prediction of Oncofetal miRNA Targets
2.6. Assessment of Oncofetal miRNAs as Cancer Diagnostic Markers
2.7. Association of Oncofetal miRNAs with Survival
2.8. Software for Statistical Analysis and Illustrations
3. Results
3.1. Novel miRNA Discovery and miRNA Profiling in Lung Tissues
3.2. Differential Expression of miRNAs in Lung Adenocarcinoma
3.3. Congruence between miRNA Expression in Lung Adenocarcinoma and the Fetal Lung
3.4. Lung Oncofetal miRNAs Target Cancer-Related Pathways
3.5. Lung Oncofetal miRNAs Hold Promise as Biomarkers of Non-Small Cell Lung Cancer
3.6. Lung Oncofetal miRNA Expression Is Inversely Correlated with Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yantiss, R.K.; Woda, B.A.; Fanger, G.R.; Kalos, M.; Whalen, G.F.; Tada, H.; Andersen, D.K.; Rock, K.L.; Dresser, K. KOC (K homology domain containing protein overexpressed in cancer): A novel molecular marker that distinguishes between benign and malignant lesions of the pancreas. Am. J. Surg. Pathol. 2005, 29, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Fung, S.W.; Cheung, P.F.; Yip, C.W.; Ng, L.W.; Cheung, T.T.; Chong, C.C.; Lee, C.; Lai, P.B.; Chan, A.W.; Tsao, G.S.; et al. The ATP-binding cassette transporter ABCF1 is a hepatic oncofetal protein that promotes chemoresistance, EMT and cancer stemness in hepatocellular carcinoma. Cancer Lett. 2019, 457, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Boyerinas, B.; Park, S.M.; Shomron, N.; Hedegaard, M.M.; Vinther, J.; Andersen, J.S.; Feig, C.; Xu, J.; Burge, C.B.; Peter, M.E. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008, 68, 2587–2591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, D.; Hayakawa, K.; Ohgane, J.; Hirosawa, M.; Nakao, Y.; Tanaka, S.; Shiota, K. An epigenetic regulatory element of the Nodal gene in the mouse and human genomes. Mech. Dev. 2015, 136, 143–154. [Google Scholar] [CrossRef]
- Galle, P.R.; Foerster, F.; Kudo, M.; Chan, S.L.; Llovet, J.M.; Qin, S.; Schelman, W.R.; Chintharlapalli, S.; Abada, P.B.; Sherman, M.; et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. Off. J. Int. Assoc. Study Liver 2019, 39, 2214–2229. [Google Scholar] [CrossRef] [Green Version]
- Muller, S.; Bley, N.; Busch, B.; Glass, M.; Lederer, M.; Misiak, C.; Fuchs, T.; Wedler, A.; Haase, J.; Bertoldo, J.B.; et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 2020, 48, 8576–8590. [Google Scholar] [CrossRef]
- Chen, M.; Li, L.; Zheng, P.S. SALL4 promotes the tumorigenicity of cervical cancer cells through activation of the Wnt/beta-catenin pathway via CTNNB1. Cancer Sci. 2019, 110, 2794–2805. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Jang, J.Y.; Jeong, S.W.; Cho, Y.K.; Lee, S.H.; Kim, S.G.; Cha, S.W.; Kim, Y.S.; Cho, Y.D.; Kim, H.S.; et al. Usefulness of AFP, AFP-L3, and PIVKA-II, and their combinations in diagnosing hepatocellular carcinoma. Medicine 2017, 96, e5811. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Chen, Z.; Zeng, H.; Guo, Y.; Liu, P.; Pan, H.; Deng, A.; Hu, J. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J. Exp. Clin. Cancer Res. CR 2010, 29, 151. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.L.; Wang, H.; Liu, J.; Wang, Z.X. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol. Cell. Biochem. 2013, 372, 35–45. [Google Scholar] [CrossRef]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef] [Green Version]
- Montani, F.; Marzi, M.J.; Dezi, F.; Dama, E.; Carletti, R.M.; Bonizzi, G.; Bertolotti, R.; Bellomi, M.; Rampinelli, C.; Maisonneuve, P.; et al. miR-Test: A blood test for lung cancer early detection. J. Natl. Cancer Inst. 2015, 107, djv063. [Google Scholar] [CrossRef] [Green Version]
- Peng, F.; Li, T.T.; Wang, K.L.; Xiao, G.Q.; Wang, J.H.; Zhao, H.D.; Kang, Z.J.; Fan, W.J.; Zhu, L.L.; Li, M.; et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8, e2569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker-Santos, D.D.; Thu, K.L.; English, J.C.; Pikor, L.A.; Martinez, V.D.; Zhang, M.; Vucic, E.A.; Luk, M.T.; Carraro, A.; Korbelik, J.; et al. Developmental transcription factor NFIB is a putative target of oncofetal miRNAs and is associated with tumour aggressiveness in lung adenocarcinoma. J. Pathol. 2016, 240, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, P.; Wang, F.; Zhang, H.; Yang, Y.; Shi, C.; Xia, Y.; Peng, J.; Liu, W.; Yang, Z.; et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat. Commun. 2012, 3, 1291. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.; Kosaka, N.; Tanaka, M.; Koizumi, F.; Kanai, Y.; Mizutani, T.; Murakami, Y.; Kuroda, M.; Miyajima, A.; Kato, T.; et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2009, 14, 529–538. [Google Scholar] [CrossRef]
- Cushing, L.; Jiang, Z.; Kuang, P.; Lu, J. The roles of microRNAs and protein components of the microRNA pathway in lung development and diseases. Am. J. Respir. Cell Mol. Biol. 2015, 52, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, W.L. Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma. Available online: https://identifiers.org/geo:GSE175462 (accessed on 25 May 2021).
- Fehlmann, T.; Backes, C.; Kahraman, M.; Haas, J.; Ludwig, N.; Posch, A.E.; Wurstle, M.L.; Hubenthal, M.; Franke, A.; Meder, B.; et al. Web-based NGS data analysis using miRMaster: A large-scale meta-analysis of human miRNAs. Nucleic Acids Res. 2017, 45, 8731–8744. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, D360–D370. [Google Scholar] [CrossRef]
- Rahmati, S.; Abovsky, M.; Pastrello, C.; Jurisica, I. pathDIP: An annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucleic Acids Res. 2017, 45, D419–D426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backes, C.; Meder, B.; Hart, M.; Ludwig, N.; Leidinger, P.; Vogel, B.; Galata, V.; Roth, P.; Menegatti, J.; Grasser, F.; et al. Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res. 2016, 44, e53. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Chen, Y.; Wu, Y.; Wang, Z.; Zhou, A.; Zhang, S.; Lin, K.; Aldape, K.; Majumder, S.; Lu, Z.; et al. Tumour suppressor TRIM33 targets nuclear beta-catenin degradation. Nat. Commun. 2015, 6, 6156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, S.; Golpon, H.; Zardo, P.; Borlak, J. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. Res. J. Lab. Clin. Med. 2021, 230, 164–196. [Google Scholar] [CrossRef]
- Sharma, S.; Kho, A.T.; Chhabra, D.; Haley, K.; Vyhlidal, C.; Gaedigk, R.; Leeder, J.S.; Tantisira, K.G.; Raby, B.; Weiss, S.T. Effect of Intrauterine Smoke Exposure on microRNA-15a Expression in Human Lung Development and Subsequent Asthma Risk. Healthcare 2020, 8, 536. [Google Scholar] [CrossRef]
- Rock, L.D.; Minatel, B.C.; Marshall, E.A.; Guisier, F.; Sage, A.P.; Barros-Filho, M.C.; Stewart, G.L.; Garnis, C.; Lam, W.L. Expanding the Transcriptome of Head and Neck Squamous Cell Carcinoma Through Novel MicroRNA Discovery. Front. Oncol. 2019, 9, 1305. [Google Scholar] [CrossRef] [PubMed]
- Wake, C.; Labadorf, A.; Dumitriu, A.; Hoss, A.G.; Bregu, J.; Albrecht, K.H.; DeStefano, A.L.; Myers, R.H. Novel microRNA discovery using small RNA sequencing in post-mortem human brain. BMC Genom. 2016, 17, 776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enterina, J.R.; Enfield, K.S.S.; Anderson, C.; Marshall, E.A.; Ng, K.W.; Lam, W.L. DLK1-DIO3 imprinted locus deregulation in development, respiratory disease, and cancer. Expert Rev. Respir. Med. 2017, 11, 749–761. [Google Scholar] [CrossRef]
- Malnou, E.C.; Umlauf, D.; Mouysset, M.; Cavaille, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 2018, 9, 706. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Vallinas, M.; Rodriguez-Paredes, M.; Albrecht, M.; Sticht, C.; Stichel, D.; Gutekunst, J.; Pitea, A.; Sass, S.; Sanchez-Rivera, F.J.; Lorenzo-Bermejo, J.; et al. Epigenetically Regulated Chromosome 14q32 miRNA Cluster Induces Metastasis and Predicts Poor Prognosis in Lung Adenocarcinoma Patients. Mol. Cancer Res. MCR 2018, 16, 390–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadal, E.; Zhong, J.; Lin, J.; Reddy, R.M.; Ramnath, N.; Orringer, M.B.; Chang, A.C.; Beer, D.G.; Chen, G. A MicroRNA cluster at 14q32 drives aggressive lung adenocarcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 3107–3117. [Google Scholar] [CrossRef] [Green Version]
- Hill, K.E.; Kelly, A.D.; Kuijjer, M.L.; Barry, W.; Rattani, A.; Garbutt, C.C.; Kissick, H.; Janeway, K.; Perez-Atayde, A.; Goldsmith, J.; et al. An imprinted non-coding genomic cluster at 14q32 defines clinically relevant molecular subtypes in osteosarcoma across multiple independent datasets. J. Hematol. Oncol. 2017, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.Y.; Du, Y.; Wang, L.; Liu, X.H.; Guo, J.; Weng, X.D. MiR-543 promotes cell proliferation and metastasis of renal cell carcinoma by targeting Dickkopf 1 through the Wnt/beta-catenin signaling pathway. J. Cancer 2018, 9, 3660–3668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.L.; Chen, X.R.; Li, Y.N.; Yan, X.Y.; Sun, J.G.; He, Q.L.; Cai, F.Z. Upregulation of miR-543-3p promotes growth and stem cell-like phenotype in bladder cancer by activating the Wnt/beta-catenin signaling pathway. Int. J. Clin. Exp. Pathol. 2017, 10, 9418–9426. [Google Scholar] [PubMed]
- Shen, D.W.; Li, Y.L.; Hou, Y.J.; Xu, Z.D.; Li, Y.Z.; Chang, J.Y. MicroRNA-543 promotes cell invasion and impedes apoptosis in pituitary adenoma via activating the Wnt/beta-catenin pathway by negative regulation of Smad7. Biosci. Biotechnol. Biochem. 2019, 83, 1035–1044. [Google Scholar] [CrossRef]
- Hong, X.; Zhang, Z.; Pan, L.; Ma, W.; Zhai, X.; Gu, C.; Zhang, Y.; Bi, X.; Huang, W.; Pei, H.; et al. MicroRNA-301b promotes the proliferation and invasion of glioma cells through enhancing activation of Wnt/beta-catenin signaling via targeting Glypican-5. Eur. J. Pharmacol. 2019, 854, 39–47. [Google Scholar] [CrossRef]
- Tang, X.; Lin, J.; Wang, G.; Lu, J. MicroRNA-433-3p promotes osteoblast differentiation through targeting DKK1 expression. PLoS ONE 2017, 12, e0179860. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Liu, T.; Xiao, Y.; Li, X.; Zhu, Y.; Zhao, Y.; Bao, J.; Wu, C. Polygonatum odoratum lectin induces apoptosis and autophagy by regulation of microRNA-1290 and microRNA-15a-3p in human lung adenocarcinoma A549 cells. Int. J. Biol. Macromol. 2016, 85, 217–226. [Google Scholar] [CrossRef]
- Fan, C.; Lin, Y.; Mao, Y.; Huang, Z.; Liu, A.Y.; Ma, H.; Yu, D.; Maitikabili, A.; Xiao, H.; Zhang, C.; et al. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget 2016, 7, 21825–21839. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chen, M.; Yu, B. miR-433 suppresses tumor progression via Smad2 in non-small cell lung cancer. Pathol. Res. Pract. 2019, 215, 152591. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Yang, D.; Gong, X.; Mo, D.; Pan, S.; Xu, J. miR-1290 promotes lung adenocarcinoma cell proliferation and invasion by targeting SOCS4. Oncotarget 2018, 9, 11977–11988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Chen, B.; Cui, F.; He, X.; Wang, W.; Wang, M. Hypoxia-induced microRNA-301b regulates apoptosis by targeting Bim in lung cancer. Cell Prolif. 2016, 49, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl. Lung Cancer Res. 2016, 5, 420–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, T.; Cui, P.; Zhou, Y.; Chen, C.; Zhao, J.; Wang, H.; Guo, M.; He, Z.; Xu, L. Antisense Oligonucleotides against miR-21 Inhibit the Growth and Metastasis of Colorectal Carcinoma via the DUSP8 Pathway. Mol. Ther. Nucleic Acids 2018, 13, 244–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Characteristics | TCGA (n = 389) | BCWH (n = 63) |
|---|---|---|
| Median Age (range) | 66 (39–88) | 70 (45–86) |
| Sex | ||
| Male | 173 (44%) | 19 (30%) |
| Female | 216 (56%) | 44 (70%) |
| Smoking Status | ||
| Current | 83 (21%) | 22 (35%) |
| Former | 224 (58%) | 18 (29%) |
| Never | 64 (16%) | 23 (37%) |
| Stage | ||
| IA | 102 (26%) | 23 (37%) |
| IB | 103 (26%) | 18 (29%) |
| IIA | 43 (11%) | 2 (3%) |
| IIB | 52 (13%) | 11 (17%) |
| IIIA | 57 (15%) | 4 (6%) |
| IIIB | 6 (2%) | 1 (2%) |
| IV | 17 (4%) | 1 (2%) |
| miRNA | Genomic Location | BCWH FL Mean 1 | BCWH ANL Mean 1 | TCGA ANL Mean 1 | BCWH LUAD Mean 1 | TCGA LUAD Mean 1 |
|---|---|---|---|---|---|---|
| hsa-miR-1290 | chr1−:18897078-18897096 | 5.86 | 0.06 | 0.31 | 1.10 | 7.86 |
| hsa-miR-1343 | chr11+:34941851-34941872 | 1.45 | 0.48 | 0.20 | 1.14 | 0.64 |
| hsa-miR-301b | chr22+:21652990-21653011 | 5.82 | 0.28 | 0.10 | 5.49 | 2.13 |
| hsa-miR-3170 | chr13+:98208533-98208554 | 1.11 | 0.41 | 0.11 | 1.24 | 1.03 |
| hsa-miR-323b | chr14+:101056233-101056255 | 7.41 | 0.34 | 0.12 | 13.15 | 7.73 |
| hsa-miR-329 | chr14+:101026797-101026819 | 9.05 | 0.50 | 0.30 | 1.62 | 0.81 |
| hsa-miR-380 | chr14+:101025021-101025042 | 4.54 | 0.23 | 0.33 | 1.35 | 0.93 |
| hsa-miR-433 | chr14+:100881897-100881918 | 12.07 | 0.52 | 0.24 | 2.62 | 1.38 |
| hsa-miR-4787 | chr3+:50675093-50675114 | 1.05 | 0.27 | 0.28 | 1.88 | 0.75 |
| hsa-miR-543 | chr14+:101032033-101032054 | 6.58 | 0.33 | 0.17 | 2.28 | 0.96 |
| hsa-miR-5684 | chr19+:12787132-12787151 | 0.67 | 0.09 | 0.16 | 0.45 | 0.55 |
| hsa-miR-627 | chr15-:42199630-42199651 | 1.20 | 0.52 | 0.49 | 1.12 | 1.15 |
| hsa-miR-6516 | chr17+:77089428-77089449 | 0.72 | 0.32 | 0.26 | 1.53 | 0.55 |
| Pathway Database | Pathway 1 | p-Value | FDR |
|---|---|---|---|
| NetPath | EGFR1 | 6 × 10−10 | 9 × 10−7 |
| REACTOME | Transcriptional Regulation by TP53 | 4 × 10−10 | 1 × 10−6 |
| REACTOME | Signal Transduction | 2 × 10−9 | 2 × 10−6 |
| ACSN2 | G2_M_CHECKPOINT | 4 × 10−9 | 3 × 10−6 |
| NetPath | AndrogenReceptor | 5 × 10−9 | 3 × 10−6 |
| NetPath | Alpha6Beta4Integrin | 8 × 10−9 | 3 × 10−6 |
| KEGG | Wnt signaling | 7 × 10−9 | 3 × 10−6 |
| ACSN2 | HEDGEHOG | 1 × 10−8 | 5 × 10−6 |
| Spike | WNT signaling | 2 × 10−8 | 6 × 10−6 |
| WikiPathways | Pathways Affected in Adenoid Cystic Carcinoma | 2 × 10−8 | 6 × 10−6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohn, D.E.; Barros-Filho, M.C.; Minatel, B.C.; Pewarchuk, M.E.; Marshall, E.A.; Vucic, E.A.; Sage, A.P.; Telkar, N.; Stewart, G.L.; Jurisica, I.; et al. Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma. Cancers 2021, 13, 2686. https://doi.org/10.3390/cancers13112686
Cohn DE, Barros-Filho MC, Minatel BC, Pewarchuk ME, Marshall EA, Vucic EA, Sage AP, Telkar N, Stewart GL, Jurisica I, et al. Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma. Cancers. 2021; 13(11):2686. https://doi.org/10.3390/cancers13112686
Chicago/Turabian StyleCohn, David E., Mateus C. Barros-Filho, Brenda C. Minatel, Michelle E. Pewarchuk, Erin A. Marshall, Emily A. Vucic, Adam P. Sage, Nikita Telkar, Greg L. Stewart, Igor Jurisica, and et al. 2021. "Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma" Cancers 13, no. 11: 2686. https://doi.org/10.3390/cancers13112686
APA StyleCohn, D. E., Barros-Filho, M. C., Minatel, B. C., Pewarchuk, M. E., Marshall, E. A., Vucic, E. A., Sage, A. P., Telkar, N., Stewart, G. L., Jurisica, I., Reis, P. P., Robinson, W. P., & Lam, W. L. (2021). Reactivation of Multiple Fetal miRNAs in Lung Adenocarcinoma. Cancers, 13(11), 2686. https://doi.org/10.3390/cancers13112686