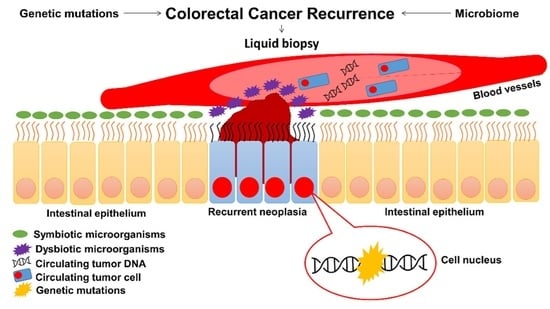
Resectable Colorectal Cancer: Current Perceptions on the Correlation of Recurrence Risk, Microbiota and Detection of Genetic Mutations in Liquid Biopsies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Genetic Mutations in CRC
2.1. Genetic Mutations Associated with CRC Recurrence
2.2. Mutations Associated with a Lower CRC Recurrence Rate
2.3. Mutations Associated with a Higher CRC Recurrence Rate
2.4. The Role of Cancer Stem Cells (CSCs) in CRC Recurrence
3. Microbiota in CRC
The Role of Gut Microbiota in CRC Recurrence
4. Liquid Biopsies: Circulating Tumor Cells and Cell-Free DNA in CRC
4.1. Association between Liquid Biopsy and CRC Recurrence
4.1.1. Circulating Tumor DNA (ctDNA)
4.1.2. Circulating Tumor Cells (CTCs)
5. Cologramme Outline
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Vyslouzil, K.; Brychtova, S.; Zboril, P.; Skalicky, P.; Vomackova, K.; Bezdekova, M.; Brychta, T. Unusual recurrent rectal carcinoma: A cancer field theory viewpoint. Biomed. Pap. 2014, 158, 433–437. [Google Scholar] [CrossRef] [Green Version]
- Kunst, N.; Alarid-Escudero, F.; Aas, E.; Coupé, V.M.H.; Schrag, D.; Kuntz, K.M. Estimating population-based recurrence rates of colorectal cancer over time in the United States. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2710–2718. [Google Scholar] [CrossRef]
- Duineveld, L.A.M.; van Asselt, K.M.; Bemelman, W.A.; Smits, A.B.; Tanis, P.J.; van Weert, H.C.P.M.; Wind, J. Symptomatic and asymptomatic colon cancer recurrence: A multicenter cohort study. Ann. Fam. Med. 2016, 14, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; He, Y.; Wang, Y.; Li, X.; Young, J.; Ioannidis, J.P.A.; Dunlop, M.G.; Theodoratou, E. Risk factors and risk prediction models for colorectal cancer metastasis and recurrence: An umbrella review of systematic reviews and meta-analyses of observational studies. BMC Med. 2020, 18, 172. [Google Scholar] [CrossRef]
- Kim, S.-E.; Paik, H.Y.; Yoon, H.; Lee, J.E.; Kim, N.; Sung, M.-K. Sex- and gender-specific disparities in colorectal cancer risk. World J. Gastroenterol. 2015, 21, 5167. [Google Scholar] [CrossRef] [PubMed]
- Arshad, H.M.S.; Tetangco, E.; Shah, N.; Kabir, C.; Raddawi, H. Racial Disparities in Colorectal Carcinoma Incidence, Severity and Survival Times Over 10 Years: A Retrospective Single Center Study. J. Clin. Med. Res. 2016, 8, 777. [Google Scholar] [CrossRef] [Green Version]
- Ohri, A.; Robinson, A.; Liu, B.; Bhuket, T.; Wong, R. Updated Assessment of Colorectal Cancer Incidence in the U.S. by Age, Sex, and Race/Ethnicity. Dig. Dis. Sci. 2020, 65, 1838–1849. [Google Scholar] [CrossRef] [PubMed]
- Lawler, M.; Alsina, D.; Adams, R.A.; Anderson, A.S.; Brown, G.; Fearnhead, N.S.; Fenwick, S.W.; Halloran, S.P.; Hochhauser, D.; Hull, M.A.; et al. Critical research gaps and recommendations to inform research prioritisation for more effective prevention and improved outcomes in colorectal cancer. Gut 2018, 67, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Genetics of Colorectal Cancer (PDQ®)–Health Professional Version-National Cancer Institute. Available online: https://www.cancer.gov/types/colorectal/hp/colorectal-genetics-pdq (accessed on 22 June 2021).
- Drew, D.A.; Devers, T.J.; O’Brien, M.J.; Horelik, N.A.; Levine, J.; Rosenberg, D.W. HD chromoendoscopy coupled with DNA mass spectrometry profiling identifies somatic mutations in microdissected human proximal aberrant crypt foci. Mol. Cancer Res. 2014, 12, 823–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebi, H.; Bando, H.; Taniguchi, H.; Sunakawa, Y.; Okugawa, Y.; Hatanaka, Y.; Hosoda, W.; Kumamoto, K.; Nakatani, K.; Yamazaki, K. Japanese Society of Medical Oncology Clinical Guidelines: Molecular Testing for Colorectal Cancer Treatment, 4th edition. Cancer Sci. 2020, 111, 3962–3969. [Google Scholar] [CrossRef]
- Afrǎsânie, V.A.; Marinca, M.V.; Alexa-Stratulat, T.; Gafton, B.; Pǎduraru, M.; Adavidoaiei, A.M.; Miron, L.; Rusu, C. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer-practical implications for the clinician. Radiol. Oncol. 2019, 53, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokota, T. Are KRAS/BRAF Mutations Potent Prognostic and/or Predictive Biomarkers in Colorectal Cancers? Anticancer. Agents Med. Chem. 2012, 12, 163–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass, A.J.; Lawrence, M.S.; Brace, L.E.; Ramos, A.H.; Drier, Y.; Cibulskis, K.; Sougnez, C.; Voet, D.; Saksena, G.; Sivachenko, A.; et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 2011, 43, 964–970. [Google Scholar] [CrossRef]
- Yu, J.; Wu, W.K.K.; Li, X.; He, J.; Li, X.X.; Ng, S.S.M.; Yu, C.; Gao, Z.; Yang, J.; Li, M.; et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 2015, 64, 636–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.K.; Kim, S.Y.; Kim, C.W.; Roh, S.A.; Ha, Y.J.; Lee, J.L.; Heo, H.; Cho, D.H.; Lee, J.S.; Kim, Y.S.; et al. A prognostic index based on an eleven gene signature to predict systemic recurrences in colorectal cancer. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef]
- Shen, Y.; Han, X.; Wang, J.; Wang, S.; Yang, H.; Lu, S.H.; Shi, Y. Prognostic impact of mutation profiling in patients with stage II and III colon cancer. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- De Sousa E Melo, F.; Colak, S.; Buikhuisen, J.; Koster, J.; Cameron, K.; De Jong, J.H.; Tuynman, J.B.; Prasetyanti, P.R.; Fessler, E.; Van Den Bergh, S.P.; et al. Methylation of cancer-stem-cell-associated wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011, 9, 476–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domingo, E.; Church, D.N.; Sieber, O.; Ramamoorthy, R.; Yanagisawa, Y.; Johnstone, E.; Davidson, B.; Kerr, D.J.; Tomlinson, I.P.M.; Midgley, R. Evaluation of PIK3CA mutation as a predictor of benefit from nonsteroidal anti-inflammatory drug therapy in colorectal cancer. J. Clin. Oncol. 2013, 31, 4297–4305. [Google Scholar] [CrossRef]
- Lan, Y.T.; Chang, S.C.; Lin, P.C.; Lin, C.C.; Lin, H.H.; Huang, S.C.; Lin, C.H.; Liang, W.Y.; Chen, W.S.; Jiang, J.K.; et al. Clinicopathological and molecular features of patients with early and late recurrence after curative surgery for colorectal cancer. Cancers 2021, 13, 1883. [Google Scholar] [CrossRef]
- Vakiani, E.; Shah, R.H.; Berger, M.F.; Makohon-Moore, A.P.; Reiter, J.G.; Ostrovnaya, I.; Attiyeh, M.A.; Cercek, A.; Shia, J.; Iacobuzio-Donahue, C.A.; et al. Local recurrences at the anastomotic area are clonally related to the primary tumor in sporadic colorectal carcinoma. Oncotarget 2017, 8, 42487–42494. [Google Scholar] [CrossRef] [Green Version]
- Schweiger, T.; Hegedüs, B.; Nikolowsky, C.; Hegedüs, Z.; Szirtes, I.; Mair, R.; Birner, P.; Döme, B.; Lang, G.; Klepetko, W.; et al. EGFR, BRAF and KRAS status in patients undergoing pulmonary metastasectomy from primary colorectal carcinoma: A prospective follow-up study. Ann. Surg. Oncol. 2014, 21, 946–954. [Google Scholar] [CrossRef]
- Sakai, N.; Furukawa, K.; Takayashiki, T.; Kuboki, S.; Takano, S.; Ohtsuka, M. Differential effects of KRAS mutational status on long-term survival according to the timing of colorectal liver metastases. BMC Cancer 2021, 21. [Google Scholar] [CrossRef] [PubMed]
- Brunsell, T.H.; Sveen, A.; Bjørnbeth, B.A.; Røsok, B.I.; Danielsen, S.A.; Brudvik, K.W.; Berg, K.C.G.; Johannessen, B.; Cengija, V.; Abildgaard, A.; et al. High Concordance and Negative Prognostic Impact of RAS/BRAF/PIK3CA Mutations in Multiple Resected Colorectal Liver Metastases. Clin. Colorectal Cancer 2020, 19, e26–e47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matteo, B.; Gaetano, P.; Delfina, T.; Riccardo, M.; Roberto, S.; Guglielmo, P.; Claudia, C.; Carmelo, L.; Carla, C.; Daris, F.; et al. Immunohistochemical evaluation of microsatellite instability in resected colorectal liver metastases: A preliminary experience. Med. Oncol. 2020, 37. [Google Scholar] [CrossRef] [PubMed]
- Gmeiner, W.H. Recent advances in our knowledge of mCRC tumor biology and genetics: A focus on targeted therapy development. Onco. Targets. Ther. 2021, 14, 2121–2130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, C.; Goel, A. Role of gut microbiota in epigenetic regulation of colorectal Cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, X.; Xu, H.; Li, S.; Lau, H.C.-H.; Chen, Q.; Zhang, B.; Zhao, L.; Chen, H.; Sung, J.J.-Y.; et al. Microbial Community Heterogeneity Within Colorectal Neoplasia and its Correlation With Colorectal Carcinogenesis. Gastroenterology 2021, 160, 2395–2408. [Google Scholar] [CrossRef]
- Schulte am Esch, J.; Windmöller, B.A.; Hanewinkel, J.; Storm, J.; Förster, C.; Wilkens, L.; Krüger, M.; Kaltschmidt, B.; Kaltschmidt, C. Isolation and Characterization of Two Novel Colorectal Cancer Cell Lines, Containing a Subpopulation with Potential Stem-Like Properties: Treatment Options by MYC/NMYC Inhibition. Cancers 2020, 12, 2582. [Google Scholar] [CrossRef] [PubMed]
- Walcher, L.; Kistenmacher, A.-K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.-R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells—Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef]
- Walker, B.S.; Zarour, L.R.; Wieghard, N.; Gallagher, A.C.; Swain, J.R.; Weinmann, S.; Lanciault, C.; Billingsley, K.; Tsikitis, V.L.; Wong, M.H. Stem Cell Marker Expression in Early Stage Colorectal Cancer is Associated with Recurrent Intestinal Neoplasia. World J. Surg. 2020, 44, 3501–3509. [Google Scholar] [CrossRef] [PubMed]
- Pisano, A.; Griñan-Lison, C.; Farace, C.; Fiorito, G.; Fenu, G.; Jiménez, G.; Scognamillo, F.; Peña-Martin, J.; Naccarati, A.; Pröll, J.; et al. The Inhibitory Role of miR-486-5p on CSC Phenotype Has Diagnostic and Prognostic Potential in Colorectal Cancer. Cancers 2020, 12, 3432. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ling, Z.; Li, L. The Intestinal Microbiota and Colorectal Cancer. Front. Immunol. 2020, 11, 3100. [Google Scholar] [CrossRef] [PubMed]
- Eaden, J.A.; Abrams, K.R.; Mayberry, J.F. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut 2001, 48, 526–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heo, G.; Lee, Y.; Im, E. Interplay between the gut microbiota and inflammatory mediators in the development of colorectal cancer. Cancers 2021, 13, 734. [Google Scholar] [CrossRef]
- Collins, R.R.J.; Patel, K.; Putnam, W.C.; Kapur, P.; Rakheja, D. Oncometabolites: A new paradigm for oncology, metabolism, and the clinical laboratory. Clin. Chem. 2017, 63, 1812–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, S.D.; Oliveira, C.S.; Azevedo-Silva, J.; Casanova, M.R.; Barreto, J.; Pereira, H.; Chaves, S.R.; Rodrigues, L.R.; Casal, M.; Côrte-Real, M.; et al. The Role of Diet Related Short-Chain Fatty Acids in Colorectal Cancer Metabolism and Survival: Prevention and Therapeutic Implications. Curr. Med. Chem. 2018, 27, 4087–4108. [Google Scholar] [CrossRef]
- Ternes, D.; Karta, J.; Tsenkova, M.; Wilmes, P.; Haan, S.; Letellier, E. Microbiome in Colorectal Cancer: How to Get from Meta-omics to Mechanism? Trends Microbiol. 2020, 28, 401–423. [Google Scholar] [CrossRef]
- Zackular, J.P.; Rogers, M.A.M.; Ruffin, M.T.; Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef] [Green Version]
- Lopez, L.R.; Bleich, R.M.; Arthur, J.C. Microbiota Effects on Carcinogenesis: Initiation, Promotion, and Progression. Annu. Rev. Med. 2021, 72, 243–261. [Google Scholar] [CrossRef]
- Dalmasso, G.; Cougnoux, A.; Delmas, J.; Darfeuille-Michaud, A.; Bonnet, R. The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes 2015, 5, 675–680. [Google Scholar] [CrossRef] [Green Version]
- Rosendahl Huber, A.; Pleguezuelos-Manzano, C.; Puschhof, J. A bacterial mutational footprint in colorectal cancer genomes. Br. J. Cancer 2021, 124, 1751–1753. [Google Scholar] [CrossRef]
- Iyadorai, T.; Mariappan, V.; Vellasamy, K.M.; Wanyiri, J.W.; Roslani, A.C.; Lee, G.K.; Sears, C.; Vadivelu, J. Prevalence and association of pks+ Escherichia coli with colorectal cancer in patients at the University Malaya Medical Centre, Malaysia. PLoS ONE 2020, 15, e0228217. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.T.; Kantilal, H.K.; Davamani, F. The mechanism of bacteroides fragilis toxin contributes to colon cancer formation. Malays. J. Med. Sci. 2020, 27, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Serna, G.; Ruiz-Pace, F.; Hernando, J.; Alonso, L.; Fasani, R.; Landolfi, S.; Comas, R.; Jimenez, J.; Elez, E.; Bullman, S.; et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020, 31, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- O’Dwyer, P.J.; Martin, E.W. Viable intraluminal tumour cells and local regional tumour growth in experimental colon cancer. Ann. R. Coll. Surg. Engl. 1989, 71, 54–56. [Google Scholar] [PubMed]
- Yu, S.Y.; Xie, Y.H.; Qiu, Y.W.; Chen, Y.X.; Fang, J.Y. Moderate alteration to gut microbiota brought by colorectal adenoma resection. J. Gastroenterol. Hepatol. 2019, 34, 1758–1765. [Google Scholar] [CrossRef]
- Shogan, B.D.; Belogortseva, N.; Luong, P.M.; Zaborin, A.; Lax, S.; Bethel, C.; Ward, M.; Muldoon, J.P.; Singer, M.; An, G.; et al. Collagen degradation and MMP9 activation by Enterococcus faecalis contribute to intestinal anastomotic leak. Sci. Transl. Med. 2015, 7. [Google Scholar] [CrossRef] [Green Version]
- Shogan, B.D.; Smith, D.P.; Christley, S.; Gilbert, J.A.; Zaborina, O.; Alverdy, J.C. Intestinal anastomotic injury alters spatially defined microbiome composition and function. Microbiome 2014, 2. [Google Scholar] [CrossRef] [Green Version]
- Olivas, A.D.; Shogan, B.D.; Valuckaite, V.; Zaborin, A.; Belogortseva, N.; Musch, M.; Meyer, F.L.; Trimble, W.; An, G.; Gilbert, J.; et al. Intestinal Tissues Induce an SNP Mutation in Pseudomonas aeruginosa That Enhances Its Virulence: Possible Role in Anastomotic Leak. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colov, E.P.; Degett, T.H.; Raskov, H.; Gögenur, I. The impact of the gut microbiota on prognosis after surgery for colorectal cancer–A systematic review and meta-analysis. APMIS 2020, 128, 162–176. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, Y.; Yu, J.; Chen, T.; Wu, Y.; Shi, L.; Li, Q.; Wu, J.; Fu, X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget 2017, 8, 31802–31814. [Google Scholar] [CrossRef]
- Rastogi, Y.R.; Saini, A.K.; Thakur, V.K.; Saini, R.V. New insights into molecular links between microbiota and gastrointestinal cancers: A literature review. Int. J. Mol. Sci. 2020, 21, 3212. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.R.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Yu, T.C.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Yang, Y.; Weng, W.; Guo, B.; Cai, G.; Ma, Y.; Cai, S. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal caner. J. Exp. Clin. Cancer Res. 2019, 38. [Google Scholar] [CrossRef]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Yi Liang, Q.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Gaines, S.; van Praagh, J.B.; Williamson, A.J.; Jacobson, R.A.; Hyoju, S.; Zaborin, A.; Mao, J.; Koo, H.Y.; Alpert, L.; Bissonnette, M.; et al. Western Diet Promotes Intestinal Colonization by Collagenolytic Microbes and Promotes Tumor Formation After Colorectal Surgery. Gastroenterology 2020, 158, 958–970.e2. [Google Scholar] [CrossRef] [PubMed]
- Gasser, M.; Lissner, R.; Nawalaniec, K.; Hsiao, L.L.; Waaga-Gasser, A.M. KMP01D Demonstrates Beneficial Anti-inflammatory Effects on Immune Cells: An ex vivo Preclinical Study of Patients With Colorectal Cancer. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Colorectal Cancer: Seed and Soil Hypothesis Revisited|Anticancer Research. Available online: https://ar.iiarjournals.org/content/34/5/2087.long (accessed on 22 June 2021).
- Huang, Q.Y.; Yao, F.; Zhou, C.R.; Huang, X.Y.; Wang, Q.; Long, H.; Wu, Q.M. Role of gut microbiome in regulating the effectiveness of metformin in reducing colorectal cancer in type 2 diabetes. World J. Clin. Cases 2020, 8, 6213–6228. [Google Scholar] [CrossRef] [PubMed]
- Bryrup, T.; Thomsen, C.W.; Kern, T.; Allin, K.H.; Brandslund, I.; Jørgensen, N.R.; Vestergaard, H.; Hansen, T.; Hansen, T.H.; Pedersen, O.; et al. Metformin-induced changes of the gut microbiota in healthy young men: Results of a non-blinded, one-armed intervention study. Diabetologia 2019, 62. [Google Scholar] [CrossRef] [Green Version]
- Demb, J.; Yaseyyedi, A.; Liu, L.; Bustamante, R.; Earles, A.; Ghosh, P.; Gutkind, J.S.; Gawron, A.J.; Kaltenbach, T.R.; Martinez, M.E.; et al. Metformin is Associated with Reduced Odds for Colorectal Cancer among Persons with Diabetes. Clin. Transl. Gastroenterol. 2019, 10. [Google Scholar] [CrossRef]
- Xi, Y.; Yuefen, P.; Wei, W.; Quan, Q.; Jing, Z.; Jiamin, X.; Shuwen, H. Analysis of prognosis, genome, microbiome, and microbial metabolome in different sites of colorectal cancer. J. Transl. Med. 2019, 17, 353. [Google Scholar] [CrossRef] [Green Version]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossettid, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeDecker, L.; Coppedge, B.; Avelar-Barragan, J.; Karnes, W.; Whiteson, K. Microbiome distinctions between the CRC carcinogenic pathways. Gut Microbes 2021, 1–12. [Google Scholar] [CrossRef]
- Kim, K.; Castro, E.J.T.; Shim, H.; Advincula, J.V.G.; Kim, Y.W. Differences regarding the molecular features and gut microbiota between right and left colon cancer. Ann. Coloproctol. 2018, 34, 292–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fong, W.; Li, Q.; Yu, J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef] [PubMed]
- Pearson, T.; Caporaso, J.G.; Yellowhair, M.; Bokulich, N.A.; Padi, M.; Roe, D.J.; Wertheim, B.C.; Linhart, M.; Martinez, J.A.; Bilagody, C.; et al. Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med. 2019, 8, 617–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boursi, B.; Mamtani, R.; Haynes, K.; Yang, Y.X. Recurrent antibiotic exposure may promote cancer formation-Another step in understanding the role of the human microbiota? Eur. J. Cancer 2015, 51, 2655–2664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phipps, O.; Al-Hassi, H.O.; Quraishi, M.N.; Dickson, E.A.; Segal, J.; Steed, H.; Kumar, A.; Acheson, A.G.; Beggs, A.D.; Brookes, M.J. Oral and intravenous iron therapy differentially alter the on-and off-tumor microbiota in anemic colorectal cancer patients. Cancers 2021, 13, 1341. [Google Scholar] [CrossRef]
- Zhang, X.; Browman, G.; Siu, W.; Basen-Engquist, K.M.; Hanash, S.M.; Hoffman, K.L.; Okhuysen, P.C.; Scheet, P.; Petrosino, J.F.; Kopetz, S.; et al. The BE GONE trial study protocol: A randomized crossover dietary intervention of dry beans targeting the gut microbiome of overweight and obese patients with a history of colorectal polyps or cancer. BMC Cancer 2019, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Food and Microbiome Longitudinal Investigation (FAMiLI). Available online: https://clinicaltrials.gov/ct2/show/NCT03293758 (accessed on 22 June 2021).
- Gut Microbiome in Fecal Samples from Patients with Metastatic Cancer Undergoing Chemotherapy or Immunotherapy. Available online: https://clinicaltrials.gov/ct2/show/NCT02960282 (accessed on 22 June 2021).
- Metagenomic Evaluation of the Gut Microbiome in Patients with Lynch Syndrome and Other Hereditary Colonic Polyposis Syndromes. Available online: https://clinicaltrials.gov/ct2/show/NCT02371135 (accessed on 22 June 2021).
- Metabiomics Colon Cancer Clinical Research Study. Available online: https://clinicaltrials.gov/ct2/show/NCT02151123 (accessed on 22 June 2021).
- Krebs, M.G.; Hou, J.M.; Ward, T.H.; Blackhall, F.H.; Dive, C. Circulating tumour cells: Their utility in cancer management and predicting outcomes. Ther. Adv. Med. Oncol. 2010, 2, 351–365. [Google Scholar] [CrossRef] [Green Version]
- Fiala, C.; Diamandis, E.P. New approaches for detecting cancer with circulating cell-free DNA. BMC Med. 2019, 17, 159. [Google Scholar] [CrossRef] [Green Version]
- Vietsch, E.E.; Wellstein, A. Circulating DNA in cancer diagnosis and prognosis. In Oncogenomics: From Basic Research to Precision Medicine; Elsevier: Amsterdam, The Netherlands, 2018; pp. 207–218. ISBN 9780128117859. [Google Scholar]
- Konczalla, L.; Wöstemeier, A.; Kemper, M.; Karstens, K.F.; Izbicki, J.; Reeh, M. Clinical significance of circulating tumor cells in gastrointestinal carcinomas. Diagnostics 2020, 10, 192. [Google Scholar] [CrossRef] [Green Version]
- Takeda, K.; Yamada, T.; Takahashi, G.; Iwai, T.; Ueda, K.; Kuriyama, S.; Koizumi, M.; Matsuda, A.; Shinji, S.; Ohta, R.; et al. Analysis of colorectal cancer-related mutations by liquid biopsy: Utility of circulating cell-free DNA and circulating tumor cells. Cancer Sci. 2019, 110, 3497–3509. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef]
- Bi, F.; Wang, Q.; Dong, Q.; Wang, Y.; Zhang, L.; Zhang, J. Circulating tumor DNA in colorectal cancer: Opportunities and challenges. Am. J. Transl. Res. 2020, 12, 1044–1055. [Google Scholar]
- Vidal, J.; Muinelo, L.; Dalmases, A.; Jones, F.; Edelstein, D.; Iglesias, M.; Orrillo, M.; Abalo, A.; Rodríguez, C.; Brozos, E.; et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann. Oncol. 2017, 28, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Schøler, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- Wills, B.; Gorse, E.; Lee, V. Role of liquid biopsies in colorectal cancer. Curr. Probl. Cancer 2018, 42, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhao, H. Liquid biopsy in tumors: Opportunities and challenges. Ann. Transl. Med. 2018, 6, S89. [Google Scholar] [CrossRef] [PubMed]
- Sisson, B.A.; Uvalic, J.; Kelly, K.; Selvam, P.; Hesse, A.N.; Ananda, G.; Chandok, H.; Bergeron, D.; Holinka, L.; Reddi, H.V. Technical and Regulatory Considerations for Taking Liquid Biopsy to the Clinic: Validation of the JAX PlasmaMonitor TM Assay. Biomark. Insights 2019, 14, 117727191982654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freidin, M.B.; Freydina, D.V.; Leung, M.; Fernandez, A.M.; Nicholson, A.G.; Lim, E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin. Chem. 2015, 61, 1299–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loaiza-Bonilla, A.; Benson, A.B.; Grothey, A.; Karimi, M.; Klempner, S.J.; Lin, D.; Mahtani, R.; Soares, H.P. Use of Molecular Assays and Circulating Tumor DNA in Early-Stage Colorectal Cancer: A Roundtable Discussion of the Gastrointestinal Cancer Therapy Expert Group. Oncologist 2021, 25, 1–9. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yokoyama, S.; Matsuda, K.; Tamura, K.; Mitani, Y.; Iwamoto, H.; Mizumoto, Y.; Murakami, D.; Kitahata, Y.; Yamaue, H. Preoperative detection of KRAS mutated circulating tumor DNA is an independent risk factor for recurrence in colorectal cancer. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Boysen, A.K.; Pallisgaard, N.; Andersen, C.S.A.; Spindler, K.L.G. Circulating tumor DNA as a marker of minimal residual disease following local treatment of metastases from colorectal cancer. Acta Oncol. 2020, 59, 1424–1429. [Google Scholar] [CrossRef]
- Symonds, E.L.; Pedersen, S.K.; Murray, D.; Byrne, S.E.; Roy, A.; Karapetis, C.; Hollington, P.; Rabbitt, P.; Jones, F.S.; LaPointe, L.; et al. Circulating epigenetic biomarkers for detection of recurrent colorectal cancer. Cancer 2020, 126, 1460–1469. [Google Scholar] [CrossRef]
- Benešová, L.; Hálková, T.; Ptáčková, R.; Semyakina, A.; Menclová, K.; Pudil, J.; Ryska, M.; Levý, M.; Šimša, J.; Pazdírek, F.; et al. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J. Gastroenterol. 2019, 25, 6939–6948. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating tumor dna analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019, 5, 1710–1717. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients with Stages i to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yukami, H.; Saori, M.; Kotani, D.; Oki, E.; Taniguchi, H.; Nakamura, Y.; Kato, T.; Takemasa, I.; Yamanaka, T.; Shirasu, H.; et al. 113TiP Prospective observational study monitoring circulating tumour DNA in resectable colorectal cancer patients undergoing radical surgery: GALAXY study in CIRCULATE-Japan. Ann. Oncol. 2020, 31, S1285–S1286. [Google Scholar] [CrossRef]
- Yukami, H.; Mishima, S.; Kotani, D.; Oki, E.; Taniguchi, H.; Nakamura, Y.; Kato, T.; Takemasa, I.; Yamanaka, T.; Shirasu, H.; et al. P-120 Prospective observational study monitoring circulating tumor DNA in resectable colorectal cancer patients undergoing radical surgery: GALAXY study in CIRCULATE-Japan (trial in progress). Ann. Oncol. 2020, 31, S128–S129. [Google Scholar] [CrossRef]
- |NIPH Clinical Trials Search. Available online: https://rctportal.niph.go.jp/en/detail?trial_id=JapicCTI-205363 (accessed on 22 June 2021).
- BESPOKE Study of ctDNA Guided Therapy in Colorectal Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT04264702 (accessed on 22 June 2021).
- Schraa, S.J.; Van Rooijen, K.L.; Van Der Kruijssen, D.E.W.; Rubio Alarcón, C.; Phallen, J.; Sausen, M.; Simmons, J.; Coupé, V.M.H.; Van Grevenstein, W.M.U.; Elias, S.; et al. Circulating tumor DNA guided adjuvant chemotherapy in stage II colon cancer (MEDOCC-CrEATE): Study protocol for a trial within a cohort study. BMC Cancer 2020, 20. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Xie, H.; Urrutia, R.; Mahipal, A. The promise of circulating tumor DNA (ctDNA) in the management of early-stage colon cancer: A critical review. Cancers 2020, 12, 2808. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wei, C.; Wang, S.; Han, S.; Shi, D.; Zhang, C.; Lin, X.; Dou, R.; Xiong, B. Combined features based on preoperative controlling nutritional status score and circulating tumour cell status predict prognosis for colorectal cancer patients treated with curative resection. Int. J. Biol. Sci. 2019, 15, 1325–1335. [Google Scholar] [CrossRef] [Green Version]
- Arrazubi, V.; Mata, E.; Antelo, M.L.; Tarifa, A.; Herrera, J.; Zazpe, C.; Teijeira, L.; Viudez, A.; Suárez, J.; Hernández, I.; et al. Circulating Tumor Cells in Patients Undergoing Resection of Colorectal Cancer Liver Metastases. Clinical Utility for Long-Term Outcome: A Prospective Trial. Ann. Surg. Oncol. 2019, 26, 2805–2811. [Google Scholar] [CrossRef]
- Yang, C.; Shi, D.; Wang, S.; Wei, C.; Zhang, C.; Xiong, B. Prognostic value of pre-and post-operative circulating tumor cells detection in colorectal cancer patients treated with curative resection: A prospective cohort study based on iset device. Cancer Manag. Res. 2018, 10, 4135–4144. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.T.; Huang, M.Y.; Yeh, Y.S.; Huang, C.W.; Tsai, H.L.; Cheng, T.L.; Wang, J.Y. A prospective study of comparing multi-gene biomarker chip and serum carcinoembryonic antigen in the postoperative surveillance for patients with stage I-III colorectal cancer. PLoS ONE 2016, 11, e0163264. [Google Scholar] [CrossRef] [PubMed]
- Tseng, J.Y.; Yang, C.Y.; Yang, S.H.; Lin, J.K.; Lin, C.H.; Jiang, J.K. Circulating CD133+/ESA+ cells in colorectal cancer patients. J. Surg. Res. 2015, 199, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Galizia, G.; Gemei, M.; Orditura, M.; Romano, C.; Zamboli, A.; Castellano, P.; Mabilia, A.; Auricchio, A.; de Vita, F.; Del Vecchio, L.; et al. Postoperative Detection of Circulating Tumor Cells Predicts Tumor Recurrence in Colorectal Cancer Patients. J. Gastrointest. Surg. 2013, 17, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, N.N.; Reissfelder, C.; Mühlbayer, M.; Weidmann, K.; Kahlert, C.; Büchler, M.W.; Weitz, J.; Koch, M. Correlation of circulating angiogenic factors with circulating tumor cells and disease recurrence in patients undergoing curative resection for colorectal liver metastases. Ann. Surg. Oncol. 2011, 18, 2182–2191. [Google Scholar] [CrossRef]
- Lu, C.Y.; Uen, Y.H.; Tsai, H.L.; Chuang, S.C.; Hou, M.F.; Wu, D.C.; Hank Juo, S.H.; Lin, S.R.; Wang, J.Y. Molecular detection of persistent postoperative circulating tumour cells in stages II and III colon cancer patients via multiple blood sampling: Prognostic significance of detection for early relapse. Br. J. Cancer 2011, 104, 1178–1184. [Google Scholar] [CrossRef] [Green Version]
- Vardakis, N.; Messaritakis, I.; Papadaki, C.; Agoglossakis, G.; Sfakianaki, M.; Saridaki, Z.; Apostolaki, S.; Koutroubakis, I.; Perraki, M.; Hatzidaki, D.; et al. Prognostic significance of the detection of peripheral blood CEACAM5mRNA-positive cells by real-time polymerase chain reaction in operable colorectal cancer. Clin. Cancer Res. 2011, 17. [Google Scholar] [CrossRef] [Green Version]
- Messaritakis, I.; Sfakianaki, M.; Papadaki, C.; Koulouridi, A.; Vardakis, N.; Koinis, F.; Hatzidaki, D.; Georgoulia, N.; Kladi, A.; Kotsakis, A.; et al. Prognostic significance of CEACAM5mRNA-positive circulating tumor cells in patients with metastatic colorectal cancer. Cancer Chemother. Pharmacol. 2018, 82, 767–775. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yoshino, T. Clinical Utility of Analyzing Circulating Tumor DNA in Patients with Metastatic Colorectal Cancer. Oncologist 2018, 23, 1310–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.C.; Rhim, A.D.; Manning, S.L.; Brennan, L.; Mansour, A.I.; Rustgi, A.K.; Damjanov, N.; Troxel, A.B.; Rickels, M.R.; Ky, B.; et al. Effects of exercise on circulating tumor cells among patients with resected stage I-III colon cancer. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [Green Version]
- GIC SG-GastroIntestinal Cancer Study Group. Available online: https://www.emkapes.gr/ (accessed on 22 June 2021).
- West, N.P.; Hohenberger, W.; Weber, K.; Perrakis, A.; Finan, P.J.; Quirke, P. Complete mesocolic excision with central vascular ligation produces an oncologically superior specimen compared with standard surgery for carcinoma of the colon. J. Clin. Oncol. 2010, 28, 272–278. [Google Scholar] [CrossRef] [Green Version]
- Nagtegaal, I.D.; Van de Velde, C.J.H.; Van Der Worp, E.; Kapiteijn, E.; Quirke, P.; Van Krieken, J.H.J.M. Macroscopic evaluation of rectal cancer resection specimen: Clinical significance of the pathologist in quality control. J. Clin. Oncol. 2002, 20, 1729–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Wood, L.D.; Parsons, D.W.; Jones, S.; Lin, J.; Sjöblom, T.; Leary, R.J.; Shen, D.; Boca, S.M.; Barber, T.; Ptak, J.; et al. The genomic landscapes of human breast and colorectal cancers. Science 2007, 318, 1108–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Sargent, D.J. Molecular pathways: Microsatellite instability in colorectal cancer: Prognostic, predictive, and therapeutic implications. Clin. Cancer Res. 2012, 18, 1506–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [Green Version]
- Alves dos Santos, K.; Clemente dos Santos, I.C.; Santos Silva, C.; Gomes Ribeiro, H.; de Farias Domingos, I.; Nogueira Silbiger, V. Circulating Exosomal miRNAs as Biomarkers for the Diagnosis and Prognosis of Colorectal Cancer. Int. J. Mol. Sci. 2020, 22, 346. [Google Scholar] [CrossRef]


| Mutation | Recurrence | Method of Detection | Sample Size | Study | |
|---|---|---|---|---|---|
| LOW RISK | CDH10, COL6A3. SMAD4, TMEM132D, VCAN | mOS (80.4 m vs. 42.4 m) HR = 0.22; 95%CI (0.07–0.70); p = 0.0051 | Exome sequencing and targeted capture sequencing | 182 | ASIAN COHORT Yu et al. (2014) |
| AK2, CDC25A, HSPB1, BID, EIF4A2, ITGB1, MAP4K4, MMP12, RHOC, PTGES3, TERF2IP | HR = 1.812, 95% CI = 1.342–2.448, p < 0.001 | Transcriptomic profiling using RNA-sequencing data | 130 | CIT COHORT Kim et al. (2019) | |
| HIGH RISK | PIK3CA | Tumor recurrence p = 0.031 and poor OS (p = 0.044) | Sanger sequencing | 228 | Shen et al. (2016) |
| APC | p = 0.023; 95% CI = 0.237–0.898 | MassArray method | 1227 | Lan et al. (2021) | |
| BRAF | Multivariate analysis of OS 95%CI (1.398–6.186); p = 0.004 | MassArray method | 1227 | Lan et al. (2021) | |
| NRAS | Multivariate analysis of OS 95%CI (0.827–3.044); p = 0.005 | MassArray method | 1227 | Lan et al. (2021) | |
| KRAS | Recurrence after PM metastasectomy multivariate analysis p = 0.035 number of PMs (p = 0.037) lung as first site of recurrence after metastasectomy (p = 0.047) | Restriction fragment length analysis | 44 | Schweiger et al. (2013) | |
| KRAS in patients with synchronous CRLM | HR = 4.316 95%CI 1.973–9.845 p < 0.001 | Polymerase chain reaction (PCR)-based primer extension assay | 255 | Sakai et al. (2021) | |
| Mkras, KRAS/NRAS/BRAF | 3-year CSS (HR, 3.3; 95% CI, 1.6–6.5; p = 0.001 | Sanger sequencing, next-generation sequencing (NGS), and/or by droplet digital polymerase chain reaction (PCR) | 106 | Brunsell et al. (2019) |
| CTCs | CtDNA | Outcomes | Study |
|---|---|---|---|
| ctDNA quantification | Earlier prediction and identification of recurrence | Bi et al. (2010) | |
| Persistent post-operative CTCs in stage II/III colon cancer patients. | Strongly correlated with early relapse (p < 0.001; HR, 11.035; 95% CI: 4.396–32.190). | Lu et al. (2011) | |
| CEACAM5mRNA-positive cells, in patients with resectable CRC. | Adverse prognostic factor associated with poor outcomes. | Vardakis et al. (2011) | |
| CTCs in mesenteric circulation. | Indicators of recurrence. | Tseng et al. (2015) | |
| CTCs vs. ctDNA. | ctDNA as a preferential specimen type for mutation screening in thoracic malignancies vs. CTC DNA. | Bi et al. (2015) | |
| CTCs detection in stage I–III CRC patients after curative resection. | Significantly higher specificity, positive and negative predictive values, and accuracy for recurrence than CEA levels. | Chang et al. (2016) | |
| Inferior to CAF in recurrence prediction. | Rahbari et al. (2011) | ||
| Patients with ≥1 CTCs following stage I-III CRC resection. | Exercise was associated with a decrease in CTCs. | Brown et al. (2018) | |
| Peripheral blood CEACAM5mRNA-positive CTCs, in patients with mCRC, especially in patients with KRAS and BRAF mutated tumors. | Adverse prognostic factor correlated with poor clinical outcome. | Messaritakis et al. (2018) | |
| Post-operative—detection of CTCs. | Poor prognosis. | Yang et al. (2018) | |
| ctDNA-positive patients. | 40-fold more likely to experience disease recurrence than ctDNA-negative patients (HR, 43.5; 95% CI, 9.8–193.5 p < 0.001). | Reinert et al. (2019) | |
| Post-surgical ctDNA status in stage III colon cancer. | Independently associated with RFI (HR, 7.5; 95% CI, 3.5–16.1; p < 0.001). | Tie et al. (2019) | |
| ctDNA vs. imaging and elevated tumor markers in early recurrence detection in patients with mCRC. | A useful tool for early detection of disease recurrence superior to imaging. | Benesova et al. (2019) | |
| Pre-operative detection of ≥2 CTCs. | Recurrence/poor prognosis despite curative resection. | Arazzubi et al. (2019) | |
| ctDNA status in stage II–IV CRC who will undergo radical surgery. | Ongoing. | ❖ GALAXY, Yukami et al. (2020) | |
| Observation vs. adjuvant CAPOX in high-risk stage II or low-risk stage III colon cancer, whose ctDNA status is negative post-operatively. | Ongoing. | VEGA, phase III (2020) | |
| Trifluridine/tipiracil vs. placebo in patients with positive ctDNA status post-resection. | Ongoing. | ALTAIR double-blind, phase III (2020) | |
| Post-resection ctDNA status and OS in CRC II or III. | Ongoing. | BESPOKE CRC prospective. (2020) | |
| Effectiveness of adjuvant chemotherapy based on ctDNA status in stage II CRC. | Ongoing. | MEDOCC-CrEATE, Schraa et al. (2020) | |
| ctDNA vs. CEA. | ctDNA showed higher sensitivity over CEA and consists of an independent predictive factor of recurrence. | Symonds et al. (2020) | |
| BRAF, NRAS and KRAS mutated ctDNA. | ctDNA following local treatment of mCRC is associated with an increased risk of recurrence and a short time to failure. | Boysen et al. (2020) | |
| KRAS mutated ctDNA. | Preoperative detection of KRAS mutated ctDNA was an independent factor related to both RFI (HR = 3.08; p = 0.012) and RFS (HR = 2.18; p = 0.044). | Nakamura et al. (2021) | |
| High CTC score. | Poor prognosis/high recurrence risk in patients with stage III CRC but not in patients with stage II CRC. | Yang et al. (2021) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koulouris, A.; Tsagkaris, C.; Messaritakis, I.; Gouvas, N.; Sfakianaki, M.; Trypaki, M.; Spyrou, V.; Christodoulakis, M.; Athanasakis, E.; Xynos, E.; et al. Resectable Colorectal Cancer: Current Perceptions on the Correlation of Recurrence Risk, Microbiota and Detection of Genetic Mutations in Liquid Biopsies. Cancers 2021, 13, 3522. https://doi.org/10.3390/cancers13143522
Koulouris A, Tsagkaris C, Messaritakis I, Gouvas N, Sfakianaki M, Trypaki M, Spyrou V, Christodoulakis M, Athanasakis E, Xynos E, et al. Resectable Colorectal Cancer: Current Perceptions on the Correlation of Recurrence Risk, Microbiota and Detection of Genetic Mutations in Liquid Biopsies. Cancers. 2021; 13(14):3522. https://doi.org/10.3390/cancers13143522
Chicago/Turabian StyleKoulouris, Andreas, Christos Tsagkaris, Ippokratis Messaritakis, Nikolaos Gouvas, Maria Sfakianaki, Maria Trypaki, Vasiliki Spyrou, Manousos Christodoulakis, Elias Athanasakis, Evangelos Xynos, and et al. 2021. "Resectable Colorectal Cancer: Current Perceptions on the Correlation of Recurrence Risk, Microbiota and Detection of Genetic Mutations in Liquid Biopsies" Cancers 13, no. 14: 3522. https://doi.org/10.3390/cancers13143522
APA StyleKoulouris, A., Tsagkaris, C., Messaritakis, I., Gouvas, N., Sfakianaki, M., Trypaki, M., Spyrou, V., Christodoulakis, M., Athanasakis, E., Xynos, E., Tzardi, M., Mavroudis, D., & Souglakos, J. (2021). Resectable Colorectal Cancer: Current Perceptions on the Correlation of Recurrence Risk, Microbiota and Detection of Genetic Mutations in Liquid Biopsies. Cancers, 13(14), 3522. https://doi.org/10.3390/cancers13143522