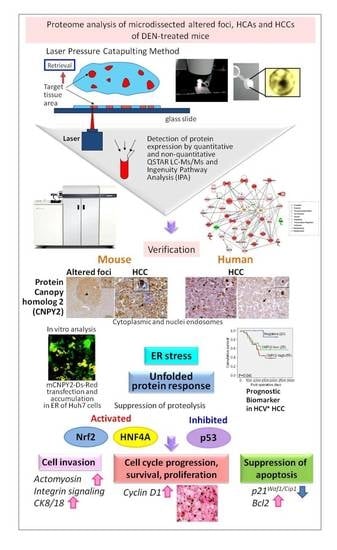
Canopy Homolog 2 as a Novel Molecular Target in Hepatocarcinogenesis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Liver Samples and Profiling by QSTAR-Elite LC-Ms/Ms
2.2. Altered Upstream Regulators and Signaling Pathways in Mice HCCs Predicted by Ingenuity Pathway Analysis (IPA)
2.3. Immunohistochemical Assessment of CNPY2 and Related Proteins in Mice Livers
2.4. In Vitro Functional Analysis of CNPY2
2.4.1. Effects of CNPY2 Knockdown with siRNAs in Huh7 and HepG2 Human Liver Cancer Cells
2.4.2. mCNPY2-Ds-Red Plasmid Transfection in Human Liver Cancer Cells
2.5. Expression of CNPY2 in HCV+ HCCs and Association with Clinicopathological Variables
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Institutional Review Board Approval
4.3. Liver Tissue from In Vivo Experiment
4.4. Laser Microdissection
4.5. Protein Identification in Microdissected Samples by QSTAR Elite LC-Ms/Ms
4.6. Ingenuity Pathway (IPA) Analysis
4.7. Immunohistochemistry and Scoring
4.8. Patients and Tissue Samples
4.9. In Vitro Experiments
4.9.1. Cell Lines and Culture Conditions
4.9.2. siRNA Knockdown of CNPY2 in Human Liver Cancer Cells
4.9.3. Generation of the Mouse CNPY2 Containing Vector
4.9.4. Transfection of CNPY2-Containing Vector in Huh7 and HepG2 Liver Cancer Cell Lines
4.9.5. WST-8 Assay
4.9.6. Invasion Assay
4.9.7. Real-Time Quantitative PCR
4.9.8. Protein Extraction and Western Blot Analysis
4.9.9. QSTAR LC-Ms/Ms and Ingenuity Pathway Analysis (IPA) of CNPY2kn Huh7 and HepG2 Cells
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, I.N.; Chen, C.H.; Sheu, J.C.; Lee, H.S.; Huang, G.T.; Yu, C.Y.; Lu, F.J.; Chow, L.P. Identification of human hepatocellular carcinoma-related biomarkers by two-dimensional difference gel electrophoresis and mass spectrometry. J. Proteome Res. 2005, 4, 2062–2069. [Google Scholar] [CrossRef]
- Lopez, L.J.; Marrero, J.A. Hepatocellular carcinoma. Curr. Opin. Gastroenterol. 2004, 20, 248–253. [Google Scholar] [CrossRef]
- Thorgeirsson, S.S.; Grisham, J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002, 31, 339–346. [Google Scholar]
- Seow, T.K.; Ong, S.E.; Liang, R.C.; Ren, E.C.; Chan, L.; Ou, K.; Chung, M.C. Two-dimensional electrophoresis map of the human hepatocellular carcinoma cell line, HCC-M, and identification of the separated proteins by mass spectrometry. Electrophoresis 2000, 21, 1787–1813. [Google Scholar] [CrossRef]
- Kakehashi, A.; Inoue, M.; Wei, M.; Fukushima, S.; Wanibuchi, H. Cytokeratin 8/18 overexpression and complex formation as an indicator of GST-P positive foci transformation into hepatocellular carcinomas. Toxicol. Appl. Pharmacol. 2009, 238, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Kakehashi, A.; Kato, A.; Inoue, M.; Ishii, N.; Okazaki, E.; Wei, M.; Tachibana, T.; Wanibuchi, H. Cytokeratin 8/18 as a new marker of mouse liver preneoplastic lesions. Toxicol. Appl. Pharmacol. 2010, 242, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Kakehashi, A.; Ishii, N.; Shibata, T.; Wei, M.; Okazaki, E.; Tachibana, T.; Fukushima, S.; Wanibuchi, H. Mitochondrial prohibitins and septin 9 are implicated in the onset of rat hepatocarcinogenesis. Toxicol. Sci. 2011, 119, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Bornhauser, B.C.; Olsson, P.A.; Lindholm, D. MSAP is a novel MIR-interacting protein that enhances neurite outgrowth and increases myosin regulatory light chain. J. Biol. Chem. 2003, 278, 35412–35420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornhauser, B.C.; Lindholm, D. MSAP enhances migration of C6 glioma cells through phosphorylation of the myosin regulatory light chain. Cell. Mol. Life Sci. 2005, 62, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Liu, B.; Wu, B.X.; Morreall, J.; Roth, B.; Davies, C.; Sun, S.; Diehl, J.A.; Li, Z. CNPY2 is a key initiator of the PERK-CHOP pathway of the unfolded protein response. Nat. Struct. Mol. Biol. 2017, 24, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Hatta, K.; Guo, J.; Ludke, A.; Dhingra, S.; Singh, K.; Huang, M.L.; Weisel, R.D.; Li, R.K. Expression of CNPY2 in mouse tissues: Quantification and localization. PLoS ONE 2014, 9, e111370. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Gong, H.; Zhai, X.; Feng, Y.; Wu, J.; He, S.; Guo, J.; Wang, X.; Guo, R.; Xie, J.; et al. Decreasing CNPY2 Expression Diminishes Colorectal Tumor Growth and Development through Activation of p53 Pathway. Am. J. Pathol. 2016, 186, 1015–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, I.B. Cancer. Addiction to oncogenes—The Achilles heal of cancer. Science 2002, 297, 63–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Z.; Fu, M.; Bouras, T.; Pestell, R.G. Signal transduction mediated by cyclin D1: From mitogens to cell proliferation: A molecular target with therapeutic potential. Cancer Treat. Res. 2004, 119, 217–237. [Google Scholar]
- Mantena, S.K.; Sharma, S.D.; Katiyar, S.K. Berberine inhibits growth, induces G1 arrest and apoptosis in human epidermoid carcinoma A431 cells by regulating Cdki-Cdk-cyclin cascade, disruption of mitochondrial membrane potential and cleavage of caspase 3 and PARP. Carcinogenesis 2006, 27, 2018–2027. [Google Scholar] [CrossRef]
- Hosseini, S.; Chamani, J.; Sinichi, M.; Bonakdar, A.M.; Azad, Z.; Ahangari, N.; Rahimi, H.R. The effect of nanomicelle curcumin, sorafenib, and combination of the two on the cyclin D1 gene expression of the hepatocellular carcinoma cell line (HUH7). Iran. J. Basic Med. Sci. 2019, 22, 1198–1202. [Google Scholar]
- Wang, N.; Wang, X.; Tan, H.Y.; Li, S.; Tsang, C.M.; Tsao, S.W.; Feng, Y. Berberine Suppresses Cyclin D1 Expression through Proteasomal Degradation in Human Hepatoma Cells. Int. J. Mol. Sci. 2016, 17, 1899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Mihic, A.; Wu, J.; Zhang, Y.; Singh, K.; Dhingra, S.; Weisel, R.D.; Li, R.K. Canopy 2 attenuates the transition from compensatory hypertrophy to dilated heart failure in hypertrophic cardiomyopathy. Eur. Heart J. 2015, 36, 2530–2540. [Google Scholar] [CrossRef]
- Ayabe, H.; Ikeda, S.; Maruyama, S.; Shioyama, S.; Kikuchi, M.; Kawaguchi, A.; Yamada, T.; Ikeda, T. Development of an efficient genotyping method to detect obese mutation in the mouse leptin gene for use in SPF barrier facilities. J. Vet. Med. Sci. 2013, 75, 633–638. [Google Scholar] [CrossRef] [Green Version]
- Pyrzynska, B.; Pilecka, I.; Miaczynska, M. Endocytic proteins in the regulation of nuclear signaling, transcription and tumorigenesis. Mol. Oncol. 2009, 3, 321–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.Z.; Wu, Z.Y.; Wang, S.H.; Ji, X.; Yang, C.X.; Xu, X.E.; Liao, L.D.; Wu, J.Y.; Li, E.M.; Zhang, K.; et al. A decision tree-based combination of ezrin-interacting proteins to estimate the prognostic risk of patients with esophageal squamous cell carcinoma. Hum. Pathol. 2017, 66, 115–125. [Google Scholar] [CrossRef]
- Peng, J.; Ou, Q.; Guo, J.; Pan, Z.; Zhang, R.; Wu, X.; Zhao, Y.; Deng, Y.; Li, C.; Wang, F.; et al. Expression of a novel CNPY2 isoform in colorectal cancer and its association with oncologic prognosis. Aging 2017, 9, 2334–2351. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Yang, P.; Aubry, M.C.; Kosari, F.; Endo, C.; Molina, J.; Vasmatzis, G. Can gene expression profiling predict survival for patients with squamous cell carcinoma of the lung? Mol. Cancer 2004, 3, 35. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, H.; Ito, S.; Ueda, T.; Morioka, Y.; Kayukawa, N.; Ueno, A.; Nakagawa, H.; Fujihara, A.; Ushijima, S.; Kanazawa, M.; et al. CNPY2 promoted the proliferation of renal cell carcinoma cells and increased the expression of TP53. BioChem. Biophys. Res. Commun. 2017, 485, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Banks, R.E.; Dunn, M.J.; Forbes, M.A.; Stanley, A.; Pappin, D.; Naven, T.; Gough, M.; Harnden, P.; Selby, P.J. The potential use of laser capture microdissection to selectively obtain distinct populations of cells for proteomic analysis—Preliminary findings. Electrophoresis 1999, 20, 689–700. [Google Scholar] [CrossRef]
- Lehmann, U.; Kreipe, H. Laser-assisted microdissection and isolation of DNA and RNA. Methods Mol. Med. 2006, 120, 65–75. [Google Scholar]
- Gluckmann, M.; Fella, K.; Waidelich, D.; Merkel, D.; Kruft, V.; Kramer, P.J.; Walter, Y.; Hellmann, J.; Karas, M.; Kroger, M. Prevalidation of potential protein biomarkers in toxicology using iTRAQ reagent technology. Proteomics 2007, 7, 1564–1574. [Google Scholar] [CrossRef]
- Tachibana, T.; Sakaguchi, N.; Miyamoto, Y.; Sekimoto, T.; Yoneda, Y.; Azuma, M. Generation and characterization of a monoclonal antibody against NPI-1 subfamily of importin alpha. Hybridoma 2008, 27, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Vauthey, J.N.; Lauwers, G.Y.; Esnaola, N.F.; Do, K.A.; Belghiti, J.; Mirza, N.; Curley, S.A.; Ellis, L.M.; Regimbeau, J.M.; Rashid, A.; et al. Simplified staging for hepatocellular carcinoma. J. Clin. Oncol. 2002, 20, 1527–1536. [Google Scholar] [CrossRef]
- Minagawa, M.; Ikai, I.; Matsuyama, Y.; Yamaoka, Y.; Makuuchi, M. Staging of hepatocellular carcinoma: Assessment of the Japanese TNM and AJCC/UICC TNM systems in a cohort of 13,772 patients in Japan. Ann. Surg. 2007, 245, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Wei, M.; Yamano, S.; Kakehashi, A.; Tamada, S.; Nakatani, T.; Wanibuchi, H. DDX39 acts as a suppressor of invasion for bladder cancer. Cancer Sci. 2012, 103, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Kakehashi, A.; Hagiwara, A.; Imai, N.; Nagano, K.; Nishimaki, F.; Banton, M.; Wei, M.; Fukushima, S.; Wanibuchi, H. Mode of action of ethyl tertiary-butyl ether hepatotumorigenicity in the rat: Evidence for a role of oxidative stress via activation of CAR, PXR and PPAR signaling pathways. Toxicol. Appl. Pharmacol. 2013, 273, 390–400. [Google Scholar] [CrossRef]




| Protein | Liver | AF | HCA | HCC |
|---|---|---|---|---|
| canopy 2 homolog (zebrafish) (CNPY2) | - | ↑ | ↑ | ↑ |
| heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) (HSPA5) | 1.20 | - | - | 2.73 |
| heat shock protein 90kDa beta (Grp94), member 1 (HSP90B1) | - | - | - | 2.19 |
| fission 1 (mitochondrial outer membrane) homolog (FIS1) | - | - | - | 2.05 |
| calreticulin (CALR) | - | ↑ | ↑ | ↑ |
| prohibitin 1 (PHB1) | - | ↑ | ↑ | ↑ |
| prohibitin 2 (PHB2) | - | ↑ | ↑ | ↑ |
| YME1-like 1 (S. cerevisiae) (YME1L1) | - | ↑ | ↑ | ↑ |
| cytokeratin 18 (CK18) | 1.27 | 2.1 | 1.49 | 1.52 |
| cytokeratin 8 (CK8) | 1.21 | 2.5 | 1.54 | 1.57 |
| fibronectin 1 (FN1) | - | - | - | ↑ |
| actin, alpha 1, skeletal muscle (ACTA1) | - | - | - | ↑ |
| actin, gamma 2, smooth muscle, enteric (ACTG2) | - | - | - | ↑ |
| Rho-associated, coiled-coil containing protein kinase 2 (ROCK2) | - | - | - | ↑ |
| myosin, heavy chain 9, non-muscle (MYH9) | - | - | - | ↑ |
| septin 9 (SEPT9) | - | ↑ | ↑ | ↑ |
| talin 1 (TLN1) | - | - | - | ↑ |
| annexin A1 (ANXA1) | - | - | - | ↑ |
| S100 calcium binding protein A8 (S100A8) | - | - | - | 3.46 |
| plectin (PLEC) | - | - | - | ↑ |
| Rho GDP dissociation inhibitor (GDI) alpha (ARHGDIA) | - | - | - | ↑ |
| aldehyde dehydrogenase 3 family, member A2 (ALDH3A2) | - | - | 2.79 | 2.05 |
| flavin containing monooxygenase 5 (FMO5) | - | - | 1.73 | 2.35 |
| superoxide dismutase 1, soluble (SOD1) | - | - | - | 0.61 |
| catalase (CAT) | - | - | - | 0.79 |
| protein disulfide isomerase family A, member 6 (PDIA6) | 1.10 | - | - | 3.18 |
| prolyl 4-hydroxylase, beta polypeptide (P4HB) | - | - | - | 2.08 |
| cytochrome P450, family 1, subfamily A, polypeptide 2 (CYP1A2) | - | - | 2.37 | 1.39 |
| cytochrome P450, family 2, subfamily E, polypeptide 1 (CYP2E1) | 1.32 | - | 1.51 | 2.98 |
| cytochrome P450, family 3, subfamily A, polypeptide 11 (CYP3A11) | - | - | - | 2.83 |
| glutathione S-transferase mu 2 (GSTM2) | - | - | 4.50 | 3.50 |
| glutathione S-transferase theta 1 (GSTT1) | - | - | - | ↑ |
| carboxylesterase 1 (monocyte/macrophage serine esterase 1) (CES1) | 1.10 | 2.88 | 1.60 | 2.74 |
| UDP-glucose pyrophosphorylase 2 (UGP2) | - | - | - | 2.63 |
| glutamate-ammonia ligase (glutamine synthetase) (GLUL(GS)) | - | 2.34 | 9.73 | 5.61 |
| ornithine aminotransferase (OAT) | - | - | - | 0.28 |
| arginase, liver (ARG1) | - | - | - | 0.64 |
| argininosuccinate lyase (ASL) | 1.07 | - | 0.23 | 1.48 |
| argininosuccinate synthetase 1 (ASS1) | 0.77 | - | 0.14 | 2.46 |
| carbamoyl-phosphate synthase 1, mitochondrial (CPS1) | 1.12 | - | - | 0.38 |
| ornithine carbamoyltransferase (OTC) | 1.34 | - | - | 0.67 |
| alpha-2-HS-glycoprotein (AHSG) | - | - | - | ↑ |
| Y box binding protein 1 (YBX1) | - | ↑ | ↑ | ↑ |
| apolipoprotein A-I (APOA1) | 1.35 | ↑ | ↑ | 8.73 |
| progesterone receptor membrane component 1 (PGRMC1) | - | - | - | 2.57 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (DDX3X) | - | - | - | 2.03 |
| Name (Symbol) | ID (GI no.) | CNPY2kn Huh7/HepG2 |
|---|---|---|
| ER and Oxidative Stress Response | ||
| calnexin (CANX) | 543920 | −2.11/−1.88 |
| calreticulin (CALR) | 117501 | ↓/↓ |
| calumenin (CALU) | 5921197 | ↓/↓ |
| heat shock 70kDa protein 2 (HSPA2) | 1708307 | ↓/↓ |
| heat shock 70kDa protein 9 (mortalin) (HSPA9) | 21264428 | ↓/↓ |
| heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) (HSPA5) | 14916999 | −2.10/−3.10 |
| Tu translation elongation factor, mitoch. (TUFM) | 1706611 | ↓/↓ |
| superoxide dismutase 2, mitochondrial (SOD2) | 134665 | ↓/↓ |
| epoxide hydrolase 1, microsomal (xenobiotic) (EPHX1) | 123926 | −2.21/−1.73 |
| peroxiredoxin 1 (PRDX1) | 548453 | ↓/↓ |
| peroxiredoxin 4 (PRDX4) | 3024727 | −2.12/−1.53 |
| Cytoskeleton organization | ||
| cytokeratin 8 (CK8) | 90110027 | −2.15/−2.77 |
| cytokeratin 18 (CK18) | 125083 | ↓/↓ |
| cytokeratin 19 (CK19) | 311033484 | ↓/↓ |
| actin, beta-like 2 (ACTBL2) | 172046825 | −2.26/−2.92 |
| myristoylated alanine-rich protein kinase C substrate (MARCKS) | 76803798 | ↓/↓ |
| MARCKS-like 1 (MARCKSL1) | 1346576 | ↓/↓ |
| profilin 1 (PFN1) | 130979 | ↓/↓ |
| cofilin 1 (non-muscle)(CFL1) | 116848 | ↓/↓ |
| tropomyosin 4 (TPM4) | 530415128 | ↓/↓ |
| Others | ||
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A (DDX39A) | 61212932 | −2.18/−1.52 |
| alpha-fetoprotein (AFP) | 120042 | −2.77/−2.43 |
| Factors | CNPY2 | ||
|---|---|---|---|
| (+) (n = 80) | (−) (n = 10) | p | |
| Age | 0.260 | ||
| >65 | 59(74%) | 9(90%) | |
| ≤65 | 21(26%) | 1(10%) | |
| Gender | 0.421 | ||
| Male | 54(67%) | 8(80%) | |
| Female | 26 (33%) | 2(20%) | |
| Smoking | 0.330 | ||
| Smoker | 43(54%) | 7(70%) | |
| Non-smoker | 37(46%) | 3(30%) | |
| Drinking | 0.171 | ||
| Drinker | 30(37%) | 6(60%) | |
| Non-drinker | 50(63%) | 4(40%) | |
| Diabetes | 0.530 | ||
| (+) | 17(21%) | 3(30%) | |
| (−) | 63(79%) | 7(70%) | |
| Cirrhosis | 0.331 | ||
| Stage 1&2 | 35(44%) | 6(60%) | |
| Stage 3&4 | 45(56%) | 4(40%) | |
| Tumor size | 0.256 | ||
| <20 mm3 | 46(58%) | 7(78%) | |
| ≥20 mm3 | 33(42%) | 2(22%) | |
| AST | 0.496 | ||
| 13–33 | 20(25%) | 3(30%) | |
| >34 ng/mL | 60(75%) | 7(70%) | |
| ALT | 0.225 | ||
| 6–27 | 54(68%) | 5(50%) | |
| >28 ng/mL | 26(32%) | 5(50%) | |
| pT | 0.016 | ||
| T1 | 19(24%) | 6(60%) | |
| T2–T4 | 61(76%) | 4(40%) | |
| pM | 0.722 | ||
| (+) | 79(99%) | 0(0%) | |
| (−) | 1(1%) | 10(100%) | |
| pB | 0.722 | ||
| (+) | 79(99%) | 0(0%) | |
| (−) | 1(1%) | 10(100%) | |
| Venous invasion | 0.038 | ||
| (+) | 55(69%) | 0(0%) | |
| (−) | 25(31%) | 10(100%) | |
| Differentiation a | 0.035 | ||
| Well | 5(6%) | 3(30%) | |
| Moderate | 33(41%) | 2(20%) | |
| Poor | 42(53%) | 5(50%) | |
| Clinical Stage b | 0.016 | ||
| I | 19(24%) | 6(60%) | |
| II | 40(50%) | 1(10%) | |
| III | 18(22%) | 3(30%) | |
| IV | 3(4%) | 0(0%) | |
| im | 0.681 | ||
| (+) | 12(15%) | 2(20%) | |
| (−) | 68(85%) | 8(80%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kakehashi, A.; Suzuki, S.; Shiota, M.; Raymo, N.; Gi, M.; Tachibana, T.; Stefanov, V.; Wanibuchi, H. Canopy Homolog 2 as a Novel Molecular Target in Hepatocarcinogenesis. Cancers 2021, 13, 3613. https://doi.org/10.3390/cancers13143613
Kakehashi A, Suzuki S, Shiota M, Raymo N, Gi M, Tachibana T, Stefanov V, Wanibuchi H. Canopy Homolog 2 as a Novel Molecular Target in Hepatocarcinogenesis. Cancers. 2021; 13(14):3613. https://doi.org/10.3390/cancers13143613
Chicago/Turabian StyleKakehashi, Anna, Shugo Suzuki, Masayuki Shiota, Nina Raymo, Min Gi, Taro Tachibana, Vasily Stefanov, and Hideki Wanibuchi. 2021. "Canopy Homolog 2 as a Novel Molecular Target in Hepatocarcinogenesis" Cancers 13, no. 14: 3613. https://doi.org/10.3390/cancers13143613
APA StyleKakehashi, A., Suzuki, S., Shiota, M., Raymo, N., Gi, M., Tachibana, T., Stefanov, V., & Wanibuchi, H. (2021). Canopy Homolog 2 as a Novel Molecular Target in Hepatocarcinogenesis. Cancers, 13(14), 3613. https://doi.org/10.3390/cancers13143613