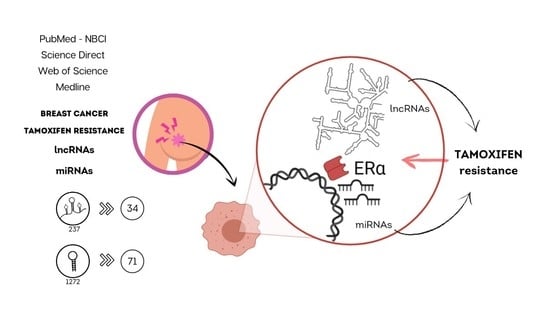
From Micro to Long: Non-Coding RNAs in Tamoxifen Resistance of Breast Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Estrogens and Estrogen Receptor Alpha
3. Mechanisms of Tamoxifen Resistance
4. Non-Coding RNAs
5. Integrative Review: Methodology
6. Results
6.1. microRNAs and Tamoxifen Resistance
| microRNA | Targets | Reference |
|---|---|---|
| Upregulated | ||
| miR-9-5p | ADIPOQ, ESR1, NCOA3 | [111,112] |
| miR-10b | HDAC4 | [56] |
| miR-15b | FOXO1 | [113] |
| miR-18a | ESR1 | [114] |
| miR-21 | PDCD4, TPM1 | [86,115,116,117] |
| miR-22 | ESR1, PTEN | [118] |
| miR-23b-3p | SLC6A14, ZBTB1 | [119,120] |
| miR-24-3p | BIM | [84] |
| miR-92a-3p | C21orf91, GPM6A, KAT2B, NEDD4L, UBE2Z, ROBO2, PCDH11X, PDS5B, TMEM87A, PRKCE | [121] |
| miR-101 | MAGI-2 | [122] |
| miR-140 | -- | [122] |
| miR-155 | SOCS6 | [123] |
| miR-181 | -- | [86] |
| miR-192-5p | ESR1 | [89] |
| miR-221/222 | P27, ESR1, TRPS1 | [86,87,88,93] |
| miR-301 | PTEN, FOXF2, COL2A1, BBC3 | [101] |
| miR-335-3p; miR-335-5p | ESR1 | [90] |
| miR-342 | -- | [124] |
| miR-342-3p; miR-342-5p | NR4A2, MAGED2, LASP1, UCP2, THSD4, PRODH, PIP4KSC, MEIS1, BMP7, PRR6, GEMIN4, GJAI, SEMA3D | [117,124] |
| miR-335-5p, miR-335-3p | ESR1 | [90] |
| miR-380 | -- | [122] |
| miR-489 | -- | [86] |
| miR-497 | -- | [125] |
| miR-519a | CDKN1A, RB1, PTEN | [102,125] |
| miR-542-5p | YWHAB, LY9, SFRP1 | [126] |
| miR-552 | -- | [122] |
| miR-575 | CDKN1B | [127] |
| miR-578 | -- | [122] |
| miR-599 | -- | [122] |
| miR-663b | TP73 | [128] |
| miR-671-3p | HNRNPA2B1 | [129] |
| miR-1266-5p | HNRNPA2B1 | [129] |
| miR-1268a | HNRNPA2B1 | [129] |
| miR-1280 | -- | [125] |
| Downregulated | ||
| let-7 | ESR1 (ER-α36 isoform) | [130] |
| miR-15a | BCL2, CCNE1 | [124,131] |
| miR-16 | BCL2, CCNE1 | [124,131] |
| miR-21 | PTEN, PDCD4 | [132] |
| miR-26a | E2F7, EZH2, ERBB22H1 | [40] |
| miR-27a;miR-27b-3p | HMGB3, NR5A2, CREB1 | [59,107,133] |
| miR-29a;miR-29b-1 | DICER1, PTEN | [104] |
| miR-29a-3p; miR-29b-3p | HNRNPA2B1, ATP5G1, ATPIF1 | [104,129] |
| miR-32-5p | CUL4B | [96] |
| miR-101 | -- | [125] |
| miR-106a | CDKN1A, PTEN, RB1 | [125] |
| miR-125a | CYP4Z1, CYP4Z2P, CDK3, HER2, HER3 | [108,134] |
| miR-125a-3p; miR-125a-5p | HDAC5 | [105,109] |
| miR-125b-5p | PAD2 | [135] |
| miR-135a | ESR1, ESRRA, NCOA1 | [94] |
| miR-146a; miR-146b | NFKB1, PI3K, TRAF6, IRAK1, PTEN | [103,117] |
| miR-148a | ALCAM | [60] |
| miR-152 | ALCAM | [60] |
| miR-181 | -- | [125] |
| miR-186-3p | EREG | [110] |
| miR-193a/b-3p | ESR1, NCOA3 | [112] |
| miR-196a | HOXB7 | [136] |
| miR-200 | ZEB1/2 | [137] |
| miR-214 | UCP2 | [58] |
| miR-222-3p | ESR1, CDKN1B, HNRNPA2B1 | [129] |
| miR-320a | ARPP19, ESRRG | [106] |
| miR-342 | TXNIP, SEMAD, BMP7, GEMIN4 | [138] |
| miR-363 | SERTAD3 | [139] |
| miR-375 | HOXB3, MTDH | [102,140] |
| miR-378a-3p | GOLT1A | [141] |
| miR-449a | ADAM22 | [142] |
| miR-451 | 14-3-3ζ | [143,144] |
| miR-484 | KLF4 | [145] |
| miR-500a-3p | LY6K | [89] |
| miR-574-3p | CLTC | [134] |
| miR-873 | CDK3 | [57,146] |
| miR-877 | -- | [134] |
| miR-4653-3p | FRS2 | [108] |
6.2. Long Non-Coding RNAs and Tamoxifen Resistance
| lncRNA | Ensembl Gene ID | Mechanism/Pathway | Reference |
|---|---|---|---|
| Upregulated | |||
| ATXN8OS | ENSG00000230223 | miR-16-5p/VASP regulation | [184] |
| BDNF-AS | ENSG00000245573 | RNH1/TRIM21/mTOR pathway | [172] |
| BLACAT1 | ENSG00000281406 | miR-503/Bcl-2 regulation | [185] |
| CCAT2 | ENSG00000280997 | Hyperactivation of ERK/MAPK pathway | [162] |
| CYTOR | ENSG00000222041 | miR-125a-5p/SRF regulation | [127] |
| DILA1 | ENST00000435697.1 | cyclin D1 binding/pRb phosphorylation | [174] |
| DSCAM-AS1 | ENSG00000235123 | Interaction with hnRNPL | [164] |
| miR-137/EPS8 regulation | [165] | ||
| FAM83H-AS1 | ENSG00000203499 | -- | [186] |
| H19 | ENSG00000130600 | Wnt/β-catenin pathway and EMT process | [150] |
| Induction of autophagy | [85] | ||
| Notch and c-Met signaling, ESR1 regulation | [149] | ||
| HNF1-AS1 | ENSG00000241388 | miR-363/SERTAD3 regulation/TGF-ß/Smad pathway regulation | [139] |
| HOTAIR | ENSG00000228630 | Promotion of ligand-independent ER activation | [166] |
| HOTAIRM1 | ENSG00000233429 | Upregulation of HOXA1 | [187] |
| LINC-ROR | ENSG00000258609 | Inhibition of autophagy | [148] |
| MAPK/ERK pathway | [147] | ||
| Epithelial-mesenchymal transition | [33] | ||
| LINP1 | ENSG00000223784 | Reduces ESR1 expression | [171] |
| LOL | ENST00000456526 | let-7 inhibition | [188] |
| MAFG-AS1 | ENSG00000265688 | miR-339-5p/CDK2 regulation | [173] |
| TMPO-AS1 | ENSG00000257167 | stabilizes ESR1 mRNA/ESR1 regulation | [189] |
| UCA1 | ENSG00000214049 | mTOR pathway | [156] |
| Feedback loop with miR-18a/HIF1A | [157] | ||
| Wnt/β-catenin pathway | [155] | ||
| PI3K/AKT pathway | [159] | ||
| -- | [157] | ||
| Downregulated | |||
| ADAMTS9-AS2 | ENSG00000241684 | miR-130a-5p/PTEN regulation | [178] |
| GAS5 | ENSG00000234741 | miR-222/PTEN regulation | [179] |
| LINC00472 | ENSG00000233237 | ERα-inducible/NF-ΚB regulation | [190] |
| LINC00894-002 | ENST00000444489 | TGF-β2 pathway | [191] |
7. Final Considerations
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Eroles, P.; Bosch, A.; Pérez-Fidalgo, J.A.; Lluch, A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat. Rev. 2012, 38, 698–707. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Breast Cancer Facts & Figures 2019–2020; American Cancer Society: Atlanta, GA, USA, 2019. [Google Scholar]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Traboulsi, T.; El Ezzy, M.; Gleason, J.; Mader, S. Antiestrogens: Structure-activity relationships and use in breast cancer treatment. J. Mol. Endocrinol. 2017, 58, R15–R31. [Google Scholar] [CrossRef] [PubMed]
- Abe, O.; Abe, R.; Enomoto, K.; Kikuchi, K.; Koyama, H.; Masuda, H.; Nomura, Y.; Sakai, K.; Sugimachi, K.; Tominaga, T.; et al. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar] [CrossRef]
- Pan, H.; Gray, R.; Braybrooke, J.; Davies, C.; Taylor, C.; McGale, P.; Peto, R.; Pritchard, K.I.; Bergh, J.; Dowsett, M.; et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 2017, 377, 1836–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ring, A.; Dowsett, M. Mechanisms of tamoxifen resistance. Endocr. Relat. Cancer 2004, 11, 643–658. [Google Scholar] [CrossRef]
- Kangaspeska, S.; Hultsch, S.; Jaiswal, A.; Edgren, H.; Mpindi, J.-P.; Eldfors, S.; Brück, O.; Aittokallio, T.; Kallioniemi, O. Systematic drug screening reveals specific vulnerabilities and co-resistance patterns in endocrine-resistant breast cancer. BMC Cancer 2016, 16, 378. [Google Scholar] [CrossRef] [Green Version]
- Hultsch, S.; Kankainen, M.; Paavolainen, L.; Kovanen, R.-M.; Ikonen, E.; Kangaspeska, S.; Pietiäinen, V.; Kallioniemi, O. Association of tamoxifen resistance and lipid reprogramming in breast cancer. BMC Cancer 2018, 18, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radhi, S. Molecular Changes During Breast Cancer and Mechanisms of Endocrine Therapy Resistance. Prog. Mol. Biol. Transl. Sci. 2016, 144, 539–562. [Google Scholar] [CrossRef]
- Sanchez, R.; Nguyen, D.; Rocha, W.; White, J.H.; Mader, S. Diversity in the mechanisms of gene regulation by estrogen receptors. BioEssays 2002, 24, 244–254. [Google Scholar] [CrossRef]
- Cotnoir-White, D.; Laperrière, D.; Mader, S. Evolution of the repertoire of nuclear receptor binding sites in genomes. Mol. Cell. Endocrinol. 2011, 334, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Brufsky, A.M.; Dickler, M.N. Estrogen Receptor-Positive Breast Cancer: Exploiting Signaling Pathways Implicated in Endocrine Resistance. Oncologist 2018, 23, 528–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begam, A.J.; Jubie, S.; Nanjan, M. Estrogen receptor agonists/antagonists in breast cancer therapy: A critical review. Bioorg. Chem. 2017, 71, 257–274. [Google Scholar] [CrossRef]
- Stender, J.D.; Frasor, J.; Komm, B.; Chang, K.C.N.; Kraus, W.L.; Katzenellenbogen, B.S. Estrogen-Regulated Gene Networks in Human Breast Cancer Cells: Involvement of E2F1 in the Regulation of Cell Proliferation. Mol. Endocrinol. 2007, 21, 2112–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourdeau, V.; Deschênes, J.; Laperrière, D.; Aid, M.; White, J.; Mader, S. Mechanisms of primary and secondary estrogen target gene regulation in breast cancer cells. Nucleic Acids Res. 2007, 36, 76–93. [Google Scholar] [CrossRef] [Green Version]
- Glück, S. Consequences of the Convergence of Multiple Alternate Pathways on the Estrogen Receptor in the Treatment of Metastatic Breast Cancer. Clin. Breast Cancer 2017, 17, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-Y.; Yin, L. Estrogen receptor alpha-36 (ER-α36): A new player in human breast cancer. Mol. Cell. Endocrinol. 2015, 418, 193–206. [Google Scholar] [CrossRef] [Green Version]
- Penot, G.; Le Péron, C.; Mérot, Y.; Grimaud-Fanouillère, E.; Ferriere, F.; Boujrad, N.; Kah, O.; Saligaut, C.; Ducouret, B.; Métivier, R.; et al. The Human Estrogen Receptor-α Isoform hERα46 Antagonizes the Proliferative Influence of hERα66 in MCF7 Breast Cancer Cells. Endocrinology 2005, 146, 5474–5484. [Google Scholar] [CrossRef] [Green Version]
- Maczis, M.A.; Maceyka, M.; Waters, M.R.; Newton, J.; Singh, M.; Rigsby, M.F.; Turner, T.; Alzubi, M.A.; Harrell, J.C.; Milstien, S.; et al. Sphingosine kinase 1 activation by estrogen receptor α36 contributes to tamoxifen resistance in breast cancer. J. Lipid Res. 2018, 59, 2297–2307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Cao, J.; Wang, Z. ER-α46, a variant of ER-α, is expressed in human breast carcinoma and enhances estrogen sensitivity in breast cancer cells. Cancer Res. 2007, 67, 986. [Google Scholar]
- Reinert, T.; Saad, E.D.; Barrios, C.H.; Bines, J. Clinical Implications of ESR1 Mutations in Hormone Receptor-Positive Advanced Breast Cancer. Front. Oncol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Katzenellenbogen, J.A.; Mayne, C.G.; Katzenellenbogen, B.S.; Greene, G.L.; Chandarlapaty, S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat. Rev. Cancer 2018, 18, 377–388. [Google Scholar] [CrossRef]
- Rondón-Lagos, M.; Villegas, V.E.; Rangel, N.; Sánchez, M.C.; Zaphiropoulos, P.G. Tamoxifen resistance: Emerging molecular targets. Int. J. Mol. Sci. 2016, 17, 1357. [Google Scholar] [CrossRef]
- Venditti, M.; Iwasiow, B.; Orr, F.W.; Shiu, R.P. C-myc gene expression alone is sufficient to confer resistance to antiestrogen in human breast cancer cells. Int. J. Cancer 2002, 99, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Wilcken, N.R.; Prall, O.W.; Musgrove, E.A.; Sutherland, R.L. Inducible overexpression of cyclin D1 in breast cancer cells reverses the growth-inhibitory effects of antiestrogens. Clin. Cancer Res. 1997, 3, 849–854. [Google Scholar] [PubMed]
- Stendahl, M.; Kronblad, Å.; Rydén, L.; Emdin, S.; Bengtsson, N.O.; Landberg, G. Cyclin D1 overexpression is a negative predictive factor for tamoxifen response in postmenopausal breast cancer patients. Br. J. Cancer 2004, 90, 1942–1948. [Google Scholar] [CrossRef] [Green Version]
- Ishii, Y.; Waxman, S.; Germain, D. Tamoxifen Stimulates the Growth of Cyclin D1–Overexpressing Breast Cancer Cells by Promoting the Activation of Signal Transducer and Activator of Transcription. Cancer Res. 2008, 68, 852–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundgren, K.; Brown, M.; Pineda, S.; Cuzick, J.; Salter, J.; Zabaglo, L.; Howell, A.; Dowsett, M.; Landberg, G. Effects of cyclin D1 gene amplification and protein expression on time to recurrence in postmenopausal breast cancer patients treated with anastrozole or tamoxifen: A TransATAC study. Breast Cancer Res. 2012, 14, R57. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Dong, N.; Wang, P.; Shi, C.; Yang, J.; Wang, J. Tamoxifen resistance and metastasis of human breast cancer cells were mediated by the membrane-associated estrogen receptor ER-α36 signaling in vitro. Cell Biol. Toxicol. 2016, 33, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, L.; Zhang, Y.; Nakata, Y.; Chan, H.L.; Morey, L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Zhang, H.-Y.; Liang, F.; Zhang, J.-W.; Wang, F.; Wang, L.; Kang, X.-G. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA. Cancer Chemother. Pharmacol. 2017, 79, 327–337. [Google Scholar] [CrossRef]
- Hull, E.E.; Montgomery, M.; Leyva, K.J. HDAC Inhibitors as Epigenetic Regulators of the Immune System: Impacts on Cancer Therapy and Inflammatory Diseases. BioMed. Res. Int. 2016, 2016, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, H.; Li, H.; Avraham, S.; Jiang, S.; Avraham, H. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor? Int. J. Cancer 2003, 107, 353–358. [Google Scholar] [CrossRef]
- Thomas, S.; Munster, P.N. Histone deacetylase inhibitor induced modulation of anti-estrogen therapy. Cancer Lett. 2009, 280, 184–191. [Google Scholar] [CrossRef] [PubMed]
- De Cremoux, P.; Dalvai, M.; N’Doye, O.; Moutahir, F.; Rolland, G.; Chouchane-Mlik, O.; Assayag, F.; Lehmann-Che, J.; Kraus-Berthie, L.; Nicolas, A.; et al. HDAC inhibition does not induce estrogen receptor in human triple-negative breast cancer cell lines and patient-derived xenografts. Breast Cancer Res. Treat. 2014, 149, 81–89. [Google Scholar] [CrossRef]
- Garmpis, N.; Christos, D.; Garmpi, A.; Kalampokas, E.; Kalampokas, T.; Spartalis, E.; Daskalopoulou, A.; Valsami, S.; Kontos, M.; Nonni, A.; et al. Histone Deacetylases as New Therapeutic Targets in Triple-negative Breast Cancer: Progress and Promises. Cancer Genom. Proteom. 2017, 14, 299–313. [Google Scholar] [CrossRef] [Green Version]
- Prossnitz, E.R.; Barton, M. The G-protein-coupled estrogen receptor GPER in health and disease. Nat. Rev. Endocrinol. 2011, 7, 715–726. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.; Ding, K.; Chong, Q.-Y.; Zhao, J.; Liu, Y.; Shao, Y.; Zhang, Y.; Yu, Q.; Xiong, Z.; Zhang, W.; et al. Post-transcriptional regulation of ERBB2 by miR26a/b and HuR confers resistance to tamoxifen in estrogen receptor-positive breast cancer cells. J. Biol. Chem. 2017, 292, 13551–13564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.-J.; Joo, H.-S.; Won, H.-Y.; Min, K.-W.; Kim, H.-Y.; Son, T.; Oh, Y.-H.; Lee, J.-Y.; Kong, G. Role of RBP2-Induced ER and IGF1R-ErbB Signaling in Tamoxifen Resistance in Breast Cancer. J. Natl. Cancer Inst. 2017, 110, 400–410. [Google Scholar] [CrossRef]
- Xu, S.; Ge, J.; Zhang, Z.; Zhou, W. MiR-129 inhibits cell proliferation and metastasis by targeting ETS1 via PI3K/AKT/mTOR pathway in prostate cancer. Biomed. Pharmacother. 2017, 96, 634–641. [Google Scholar] [CrossRef]
- Augereau, P.; Patsouris, A.; Bourbouloux, E.; Gourmelon, C.; Lacourtoisie, S.A.; Rigaud, D.B.; Soulié, P.; Frenel, J.-S.; Campone, M. Hormonoresistance in advanced breast cancer: A new revolution in endocrine therapy. Ther. Adv. Med. Oncol. 2017, 9, 335–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Du, Y.; Cui, F.; Feng, X.; Ma, Y.; Liu, H. Downregulation of BAG-1 in T47D cells promotes resistance to tamoxifen via activation of the PI3K/Akt/mTOR signaling pathway. Oncol. Rep. 2019, 41, 1901–1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Y.; Zhou, J.; Luo, X.; Chen, M.; Chen, Y.; Wang, J.; Xiong, H.; Ying, X.; Hu, W.; Zhao, W.; et al. KLF4 overcomes tamoxifen resistance by suppressing MAPK signaling pathway and predicts good prognosis in breast cancer. Cell. Signal. 2018, 42, 165–175. [Google Scholar] [CrossRef]
- Hsu, L.-H.; Chu, N.-M.; Lin, Y.-F.; Kao, S.-H. G-Protein Coupled Estrogen Receptor in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The FANTOM Consortium; Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; et al. The Transcriptional Landscape of the Mammalian Genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nat. Cell Biol. 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nat. Cell Biol. 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, T.; Gerhardt, D.J.; Dinger, M.; Crawford, J.; Trapnell, C.; A Jeddeloh, J.; Mattick, J.; Rinn, J.L. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2011, 30, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef] [PubMed]
- Antonov, I.V.; Mazurov, E.; Borodovsky, M.; A Medvedeva, Y. Prediction of lncRNAs and their interactions with nucleic acids: Benchmarking bioinformatics tools. Brief. Bioinform. 2018, 20, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Kasinski, A.; Slack, F.J. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer 2011, 11, 849–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Dong, C.; Ji, C. MicroRNA and drug resistance. Cancer Gene Ther. 2010, 17, 523–531. [Google Scholar] [CrossRef] [Green Version]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. miRNA Dysregulation in Breast Cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, A.; Ginnebaugh, K.R.; Yin, S.; Bollig-Fischer, A.; Reddy, K.B.; Sarkar, F.H. Functional role of miR-10b in tamoxifen resistance of ER-positive breast cancer cells through down-regulation of HDAC. BMC Cancer 2015, 15, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Yang, Y.; Li, H.; Leng, Y.; Qian, K.; Huang, Q.; Zhang, C.; Lu, Z.; Chen, J.; Sun, T.; et al. MiR-873 regulates ERα transcriptional activity and tamoxifen resistance via targeting CDK3 in breast cancer cells. Oncogene 2014, 34, 3895–3907. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Zou, Z.; Nie, P.; Kou, X.; Wu, B.; Wang, S.; Song, Z.; He, J. Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis. 2016, 7, e2454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.-J.; Cheng, Y.-M.; Chen, C.-C.; Chen, Y.-C.; Shen, C.-J. MiR-148a and miR-152 reduce tamoxifen resistance in ER+ breast cancer via downregulating ALCAM. Biochem. Biophys. Res. Commun. 2017, 483, 840–846. [Google Scholar] [CrossRef]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef] [Green Version]
- Gil, N.; Ulitsky, I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2019, 21, 102–117. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Mo, Y.; Peng, M.; Tang, T.; Zhong, Y.; Deng, X.; Xiong, F.; Guo, C.; Wu, X.; Li, Y.; et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 2020, 19, 1–14. [Google Scholar] [CrossRef]
- Lyle, R.; Watanabe, D.; Vruchte, D.T.; Lerchner, W.; Smrzka, O.W.; Wutz, A.; Schageman, J.; Hahner, L.; Davies, C.; Barlow, D.P. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas. Nat. Genet. 2000, 25, 19–21. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.; Kertesz, M.; Wang, J.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, T.; Dinger, M.; Sunkin, S.M.; Mehler, M.F.; Mattick, J. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Tian, H.; Yang, J.; Gong, Z. Long Noncoding RNAs Regulate Cell Growth, Proliferation, and Apoptosis. DNA Cell Biol. 2016, 35, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [Green Version]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Şenergin, H.B.; Hudson, Q.; Stocsits, R.R.; Allhoff, W.; Stricker, S.; Klement, R.M.; Warczok, K.E.; et al. Airn Transcriptional Overlap, But Not Its lncRNA Products, Induces Imprinted Igf2r Silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef]
- Santoro, F.; Mayer, D.; Klement, R.M.; Warczok, K.E.; Stukalov, A.; Barlow, D.P.; Pauler, F.M. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 2013, 140, 1184–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nat. Cell Biol. 2011, 477, 295–300. [Google Scholar] [CrossRef] [Green Version]
- Ng, S.-Y.; Johnson, R.; Stanton, L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2011, 31, 522–533. [Google Scholar] [CrossRef] [Green Version]
- Lin, N.; Chang, K.-Y.; Li, Z.; Gates, K.; Rana, Z.A.; Dang, J.; Zhang, D.; Han, T.; Yang, C.-S.; Cunningham, T.; et al. An Evolutionarily Conserved Long Noncoding RNA TUNA Controls Pluripotency and Neural Lineage Commitment. Mol. Cell 2014, 53, 1005–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.-Y.; Kuo, H.-C. The Trans-Spliced Long Noncoding RNA tsRMSTImpedes Human Embryonic Stem Cell Differentiation Through WNT5A-Mediated Inhibition of the Epithelial-to-Mesenchymal Transition. Stem Cells 2016, 34, 2052–2062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obaid, M.; Udden, S.M.N.; Deb, P.; Shihabeddin, N.; Zaki, H.; Mandal, S.S. LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages. Sci. Rep. 2018, 8, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nat. Cell Biol. 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Huang, R.; Wang, X.; Zhang, W.; Zhangyuan, G.; Jin, K.; Yu, W.; Xie, Y.; Xu, X.; Wang, H.; Sun, B. Down-Regulation of LncRNA DGCR5 Correlates with Poor Prognosis in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2016, 40, 707–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, K.; Joyce, C.E.; Buquicchio, F.; Brown, A.; Ritz, J.; Distel, R.J.; Yoon, C.H.; Novina, C.D. The lncRNA SLNCR1 Mediates Melanoma Invasion through a Conserved SRA1-like Region. Cell Rep. 2016, 15, 2025–2037. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, J.C.; Oliveira, L.C.; Mathias, C.; Pedroso, G.A.; Lemos, D.S.; Salviano-Silva, A.; Jucoski, T.S.; Lobo-Alves, S.C.; Zambalde, E.P.; Cipolla, G.A.; et al. Long non-coding RNAs in cancer: Another layer of complexity. J. Gene Med. 2019, 21, e3065. [Google Scholar] [PubMed] [Green Version]
- Huang, L.; Liang, G.; Zhang, Q.; Zhao, W. The Role of Long Noncoding RNAs in Antiestrogen Resistance in Breast Cancer: An Overview and Update. J. Breast Cancer 2020, 23, 129–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manavalan, T.T.; Teng, Y.; Appana, S.N.; Datta, S.; Kalbfleisch, T.S.; Li, Y.; Klinge, C.M. Differential expression of microRNA expression in tamoxifen-sensitive MCF-7 versus tamoxifen-resistant LY2 human breast cancer cells. Cancer Lett. 2011, 313, 26–43. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Li, Q.; Liu, C.; Wang, C.; Li, Y. Overexpression miR-24-3p repressed Bim expression to confer tamoxifen resistance in breast cancer. J. Cell. Biochem. 2019, 120, 12966–12976. [Google Scholar] [CrossRef]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 2019, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- E Miller, T.; Ghoshal, K.; Ramaswamy, B.; Roy, S.; Datta, J.; Shapiro, C.L.; Jacob, S.; Majumder, S. MicroRNA-221/222 Confers Tamoxifen Resistance in Breast Cancer by Targeting p27Kip. J. Biol. Chem. 2008, 283, 29897–29903. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.-J.; Lin, J.; Yang, H.; Kong, W.; He, L.; Ma, X.; Coppola, D.; Cheng, J.Q. MicroRNA-221/222 Negatively Regulates Estrogen Receptorα and Is Associated with Tamoxifen Resistance in Breast Cancer. J. Biol. Chem. 2008, 283, 31079–31087. [Google Scholar] [CrossRef] [Green Version]
- Han, S.-H.; Kim, H.J.; Gwak, J.M.; Kim, M.; Chung, Y.R.; Park, S.Y. MicroRNA-222 Expression as a Predictive Marker for Tumor Progression in Hormone Receptor-Positive Breast Cancer. J. Breast Cancer 2017, 20, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.S.; Park, S.J.; Lee, Y.S.; Kong, H.K.; Park, J.H. miRNAs involved in LY6K and estrogen receptor α contribute to tamoxifen-susceptibility in breast cancer. Oncotarget 2016, 7, 42261–42273. [Google Scholar] [CrossRef] [Green Version]
- Martin, E.C.; Conger, A.K.; Yan, T.J.; Hoang, V.T.; Miller, D.F.B.; Buechlein, A.; Rusch, D.B.; Nephew, K.P.; Collins-Burow, B.M.; Burow, M.E. MicroRNA-335-5p and -3p synergize to inhibit estrogen receptor alpha expression and promote tamoxifen resistance. FEBS Lett. 2016, 591, 382–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochrane, D.R.; Cittelly, D.; Howe, E.; Spoelstra, N.S.; McKinsey, E.L.; LaPara, K.; Elias, A.; Yee, D.; Richer, J.K. MicroRNAs Link Estrogen Receptor Alpha Status and Dicer Levels in Breast Cancer. Horm. Cancer 2010, 1, 306–319. [Google Scholar] [CrossRef] [Green Version]
- Di Leva, G.; Gasparini, P.; Piovan, C.; Ngankeu, A.; Garofalo, M.; Taccioli, C.; Iorio, M.; Li, M.; Volinia, S.; Alder, H.; et al. MicroRNA Cluster 221-222 and Estrogen Receptor α Interactions in Breast Cancer. J. Natl. Cancer Inst. 2010, 102, 706–721. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, M.; Chong, Q.-Y.; Zhang, M.; Zhang, X.; Hu, L.; Zhong, Y.; Qian, P.; Kong, X.J.; Tan, S.; et al. Loss of Estrogen-Regulated MIR135A1 at 3p21.1 Promotes Tamoxifen Resistance in Breast Cancer. Cancer Res. 2018, 78, 4915–4928. [Google Scholar] [CrossRef] [Green Version]
- Pagano, M.T.; Ortona, E.; Dupuis, M.L. A Role for Estrogen Receptor alpha36 in Cancer Progression. Front. Endocrinol. 2020, 11, 506. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, X.; Li, Y.; Wang, R.; Yang, Y.; Jiang, B.; Sun, G.; Shao, C.; Wang, M.; Gong, Y. CUL4B renders breast cancer cells tamoxifen-resistant via miR -32-5p/ ER -α36 axis. J. Pathol. 2021, 254, 185–198. [Google Scholar] [CrossRef]
- Shou, J.; Massarweh, S.; Osborne, C.K.; Wakeling, A.E.; Ali, S.; Weiss, H.; Schiff, R. Mechanisms of Tamoxifen Resistance: Increased Estrogen Receptor-HER2/neu Cross-Talk in ER/HER2-Positive Breast Cancer. J. Natl. Cancer Inst. 2004, 96, 926–935. [Google Scholar] [CrossRef] [Green Version]
- Musgrove, E.A.; Sutherland, R.L. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 2009, 9, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Schiff, R. Mechanisms of Endocrine Resistance in Breast Cancer. Annu. Rev. Med. 2011, 62, 233–247. [Google Scholar] [CrossRef] [Green Version]
- Milella, M.; Falcone, I.; Conciatori, F.; Incani, U.C.; Del Curatolo, A.; Inzerilli, N.; Nuzzo, C.M.A.; Vaccaro, V.; Vari, S.; Cognetti, F.; et al. PTEN: Multiple functions in human malignant tumors. Front. Oncol. 2015, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Gerster, K.; Alajez, N.M.; Tsang, J.; Waldron, L.; Pintilie, M.; Hui, A.B.; Sykes, J.; P’Ng, C.; Miller, N.; et al. MicroRNA-301 Mediates Proliferation and Invasion in Human Breast Cancer. Cancer Res. 2011, 71, 2926–2937. [Google Scholar] [CrossRef] [Green Version]
- Ward, A.; Shukla, K.; Balwierz, A.; Soons, Z.; König, R.; Şahin, Z.; Wiemann, S. MicroRNA -519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER + breast cancer. J. Pathol. 2014, 233, 368–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phuong, N.T.T.; Kim, S.K.; Im, J.H.; Yang, J.W.; Choi, M.C.; Lim, S.C.; Lee, K.Y.; Kim, Y.-M.; Yoon, J.H.; Kang, K.W. Induction of methionine adenosyltransferase 2A in tamoxifen-resistant breast cancer cells. Oncotarget 2015, 7, 13902–13916. [Google Scholar] [CrossRef]
- Muluhngwi, P.; Krishna, A.; Vittitow, S.L.; Napier, J.T.; Richardson, K.M.; Ellis, M.; Mott, J.L.; Klinge, C.M. Tamoxifen differentially regulates miR-29b-1 and miR-29a expression depending on endocrine-sensitivity in breast cancer cells. Cancer Lett. 2017, 388, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.-T.; Tsai, Y.-H.; Chen, S.-H.; Kuo, C.-W.; Kuo, Y.-L.; Lee, K.-T.; Chen, W.-C.; Wu, P.C.; Chuang, C.-Y.; Cheng, S.M.; et al. HDAC2 and HDAC5 Up-Regulations Modulate Survivin and miR-125a-5p Expressions and Promote Hormone Therapy Resistance in Estrogen Receptor Positive Breast Cancer Cells. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lü, M.; Ding, K.; Zhang, G.; Yin, M.; Yao, G.; Tian, H.; Lian, J.; Liu, L.; Liang, M.; Zhu, T.; et al. MicroRNA-320a sensitizes tamoxifen-resistant breast cancer cells to tamoxifen by targeting ARPP-19 and ERRγ*. Sci. Rep. 2015, 5, srep08735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wu, Y.; Liu, A.; Tang, X. MiR-27b is epigenetically downregulated in tamoxifen resistant breast cancer cells due to promoter methylation and regulates tamoxifen sensitivity by targeting HMGB. Biochem. Biophys. Res. Commun. 2016, 477, 768–773. [Google Scholar] [CrossRef]
- Zhong, X.; Xie, G.; Zhang, Z.; Wang, Z.; Wang, Y.; Wang, Y.; Qiu, Y.; Li, L.; Bu, H.; Li, J.; et al. MiR-4653-3p and its target gene FRS2 are prognostic biomarkers for hormone receptor positive breast cancer patients receiving tamoxifen as adjuvant endocrine therapy. Oncotarget 2016, 7, 61166–61182. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Meng, X.; Li, X.; Zhang, Y.; Li, C.; Xiang, C.; Xing, Y.; Xia, Y.; Xi, T. miR-125a-3p inhibits ERα transactivation and overrides tamoxifen resistance by targeting CDK3 in estrogen receptor–positive breast cancer. FASEB J. 2018, 32, 588–600. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Jin, Q.; Chen, C.; Liu, Y.; Ye, X.; Jiang, Y.; Ji, F.; Qian, H.; Gan, D.; Yue, S.; et al. The miR-186-3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene 2019, 38, 5551–5565. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, S.; Tang, W.; Huang, Q.; Mei, Y.; Yang, H. Exosomes from tamoxifen-resistant breast cancer cells transmit drug resistance partly by delivering miR-9-5p. Cancer Cell Int. 2021, 21, 1–15. [Google Scholar] [CrossRef]
- Pillai, M.M.; Gillen, A.; Yamamoto, T.M.; Kline, E.; Brown, J.; Flory, K.; Hesselberth, J.; Kabos, P. HITS-CLIP reveals key regulators of nuclear receptor signaling in breast cancer. Breast Cancer Res. Treat. 2014, 146, 85–97. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Li, X.; Luo, G.; Shen, M.; Shi, J.; Wang, X.; Tang, L. Paeoniflorin Sensitizes Breast Cancer Cells to Tamoxifen by Downregulating microRNA-15b via the FOXO1/CCND1/β-Catenin Axis. Drug Des. Dev. Ther. 2021, 15, 245–257. [Google Scholar] [CrossRef]
- Luengo-Gil, G.; García-Martínez, E.; Chaves-Benito, A.; Conesa-Zamora, P.; Manzano, E.N.; González-Billalabeitia, E.; García-Garre, E.; Martínez-Carrasco, A.; Vicente, V.; De La Peña, F.A. Clinical and biological impact of miR-18a expression in breast cancer after neoadjuvant chemotherapy. Cell. Oncol. 2019, 42, 627–644. [Google Scholar] [CrossRef]
- Chen, C.; Liang, Z.; Huang, W.; Li, X.; Zhou, F.; Hu, X.; Han, M.; Ding, X.; Xiang, S. Eps8 regulates cellular proliferation and migration of breast cancer. Int. J. Oncol. 2014, 46, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, R.; Shi, W.; Jiang, T.; Wang, Y.; Li, C.; Qu, X. Silencing of MicroRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT-mTOR pathway in breast cancer cells. Biomed. Pharmacother. 2016, 77, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Ye, P.; Fang, C.; Zeng, H.; Shi, Y.; Pan, Z.; An, N.; He, K.; Zhang, L.; Long, X. Differential microRNA expression profiles in tamoxifen-resistant human breast cancer cell lines induced by two methods. Oncol. Lett. 2018, 15, 3532–3539. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Zhang, X.; Han, B.; Tian, X. Sensitization to tamoxifen by tanshinone IIA in tamoxifen-resistant breast cancer cells in vitro. Inter. J. Clin. Exper. Med. 2017, 10, 2660–2671. [Google Scholar]
- Bacci, M.; Lorito, N.; Ippolito, L.; Ramazzotti, M.; Luti, S.; Romagnoli, S.; Parri, M.; Bianchini, F.; Cappellesso, F.; Virga, F.; et al. Reprogramming of Amino Acid Transporters to Support Aspartate and Glutamate Dependency Sustains Endocrine Resistance in Breast Cancer. Cell Rep. 2019, 28, 104–118.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, D.; Zhang, G.; Wu, X.; Zhou, L.; Lin, Y.; Ding, J.; An, F.; Zhan, Q. MicroRNA-23b-3p promotes pancreatic cancer cell tumorigenesis and metastasis via the JAK/PI3K and Akt/NF-κB signaling pathways. Oncol. Lett. 2020, 20, 1. [Google Scholar] [CrossRef]
- Cun, J.; Yang, Q. Bioinformatics-based interaction analysis of miR-92a-3p and key genes in tamoxifen-resistant breast cancer cells. Biomed. Pharmacother. 2018, 107, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Wu, H.; Ru, P.; Hwang, L.; Trieu, V.; Mo, Y.-Y. MicroRNA-101-mediated Akt activation and estrogen-independent growth. Oncogene 2010, 30, 822–831. [Google Scholar] [CrossRef]
- Shen, R.; Wang, Y.; Wang, C.-X.; Yin, M.; Liu, H.-L.; Chen, J.-P.; Han, J.-Q.; Wang, W.-B. MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 2015, 7, 2115–2126. [Google Scholar]
- Cittelly, D.M.; Das, P.M.; Spoelstra, N.S.; Edgerton, S.M.; Richer, J.K.; Thor, A.D.; E Jones, F. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer 2010, 9, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eskiler, G.G.; Cecener, G.; Dikmen, G.; Egeli, U.; Tunca, B. Solid lipid nanoparticles: Reversal of tamoxifen resistance in breast cancer. Eur. J. Pharm. Sci. 2018, 120, 73–88. [Google Scholar] [CrossRef]
- Zhu, Z.-M.; Liu, F.-T.; Chen, X. Low Expression of LncRNA Cancer Susceptibility Candidate 2 and its Clinical Significance in Cancer Tissues. Cell. Physiol. Biochem. 2018, 46, 1643–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, M.; Yu, H.; Piao, H. lncRNA CYTOR promotes tamoxifen resistance in breast cancer cells via sponging miR-125a-5p. Int. J. Mol. Med. 2019, 45, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Cheng, L.; Hu, P.; Liu, R. MicroRNA-663b mediates TAM resistance in breast cancer by modulating TP73 expression. Mol. Med. Rep. 2018, 18, 1120–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klinge, C.M.; Piell, K.M.; Tooley, C.S.; Rouchka, E.C. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci. Rep. 2019, 9, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Deng, C.; Lu, W.; Xiao, J.; Ma, D.; Guo, M.; Recker, R.R.; Gatalica, Z.; Wang, Z.; Xiao, G.G. let-7 MicroRNAs Induce Tamoxifen Sensitivity by Downregulation of Estrogen Receptor α Signaling in Breast Cancer. Mol. Med. 2011, 17, 1233–1241. [Google Scholar] [CrossRef]
- Chu, J.; Zhu, Y.; Liu, Y.; Sun, L.; Lv, X.; Wu, Y.; Hu, P.; Su, F.; Gong, C.; Song, E.; et al. E2F7 overexpression leads to tamoxifen resistance in breast cancer cells by competing with E2F1 at miR-15a/16 promoter. Oncotarget 2015, 6, 31944–31957. [Google Scholar] [CrossRef]
- Klinge, C.M.; Riggs, K.A.; Wickramasinghe, N.S.; Emberts, C.G.; McConda, D.B.; Barry, P.N.; Magnusen, J.E. Estrogen receptor alpha 46 is reduced in tamoxifen resistant breast cancer cells and re-expression inhibits cell proliferation and estrogen receptor alpha 66-regulated target gene transcription. Mol. Cell. Endocrinol. 2010, 323, 268–276. [Google Scholar] [CrossRef] [Green Version]
- Ljepoja, B.; García-Roman, J.; Sommer, A.-K.; Wagner, E.; Roidl, A. MiRNA-27a sensitizes breast cancer cells to treatment with Selective Estrogen Receptor Modulators. Breast 2019, 43, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Ujihira, T.; Ikeda, K.; Suzuki, T.; Yamaga, R.; Sato, W.; Horieinoue, K.; Shigekawa, T.; Osaki, A.; Saeki, T.; Okamoto, K.; et al. MicroRNA-574-3p, identified by microRNA library-based functional screening, modulates tamoxifen response in breast cancer. Sci. Rep. 2015, 5, 7641. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Miao, L.; Xue, T.; Qin, H.; Mondal, S.; Thompson, P.R.; Coonrod, S.A.; Liu, X.; Zhang, X. Inhibiting PAD2 enhances the anti-tumor effect of docetaxel in tamoxifen-resistant breast cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, K.; Park, S.; Teo, W.W.; Korangath, P.; Cho, S.; Yoshida, T.; Gyorffy, B.; Goswami, C.P.; Nakshatri, H.; Cruz, L.-A.; et al. HOXB7 Is an ERα Cofactor in the Activation of HER2 and Multiple ER Target Genes Leading to Endocrine Resistance. Cancer Discov. 2015, 5, 944–959. [Google Scholar] [CrossRef] [Green Version]
- Manavalan, T.T.; Teng, Y.; Litchfield, L.M.; Muluhngwi, P.; Al-Rayyan, N.; Klinge, C.M. Reduced Expression of miR-200 Family Members Contributes to Antiestrogen Resistance in LY2 Human Breast Cancer Cells. PLoS ONE 2013, 8, e62334. [Google Scholar] [CrossRef] [Green Version]
- Young, J.; Kawaguchi, T.; Yan, L.; Qi, Q.; Liu, S.; Takabe, K. Tamoxifen sensitivity-related microRNA-342 is a useful biomarker for breast cancer survival. Oncotarget 2017, 8, 99978–99989. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, L.; Lv, Y.; Zhang, Y.; Zhang, L.; Yu, H.; Tian, W.; Zhang, Z.; Cui, S. Silencing long non-coding RNA HNF1A-AS1 inhibits growth and resistance to TAM of breast cancer cells via the microRNA-363/SERTAD3 axis. J. Drug Target. 2021, 1–31. [Google Scholar] [CrossRef]
- Lin, Y.; Fu, F.; Chen, Y.; Qiu, W.; Lin, S.; Yang, P.; Huang, M.; Wang, C. Genetic variants in long noncoding RNA H19 contribute to the risk of breast cancer in a southeast China Han population. OncoTargets Ther. 2017, 10, 4369–4378. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Horie-Inoue, K.; Ueno, T.; Suzuki, T.; Sato, W.; Shigekawa, T.; Osaki, A.; Saeki, T.; Berezikov, E.; Mano, H.; et al. miR-378a-3p modulates tamoxifen sensitivity in breast cancer MCF-7 cells through targeting GOLT1A. Sci. Rep. 2015, 5, 13170. [Google Scholar] [CrossRef]
- Li, J.; Lu, M.; Jin, J.; Lu, X.; Xu, T.; Jin, S. miR-449a Suppresses Tamoxifen Resistance in Human Breast Cancer Cells by Targeting ADAM. Cell. Physiol. Biochem. 2018, 50, 136–149. [Google Scholar] [CrossRef]
- Bergamaschi, A.; Katzenellenbogen, B.S. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene 2011, 31, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.-R.; Song, Y.; Wan, L.-H.; Zhang, Y.-Y.; Zhou, L.-M. Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3ζ, estrogen receptor α, and autophagy. Life Sci. 2016, 149, 104–113. [Google Scholar] [CrossRef]
- Wei, Y.; Li, H.; Qu, Q. miR-484 suppresses endocrine therapy-resistant cells by inhibiting KLF4-induced cancer stem cells in estrogen receptor-positive cancers. Breast Cancer 2020, 28, 175–186. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, B.; Zhang, P.; Lian, L.; Li, L.; Qiu, Z.; Qian, K.; Chen, A.; Liu, Q.; Jiang, Y.; et al. Norcantharidin regulates ERα signaling and tamoxifen resistance via targeting miR-873/CDK3 in breast cancer cells. PLoS ONE 2019, 14, e0217181. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.-X.; Huang, J.; Yang, L.; Gong, A.-H.; Mo, Y.-Y. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Cancer 2017, 16, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jiang, B.; Zhu, H.; Qu, X.; Zhao, L.; Tan, Y.; Jiang, Y.; Liao, M.; Wu, X. Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Basak, P.; Chatterjee, S.; Bhat, V.; Su, A.; Jin, H.; Lee-Wing, V.; Liu, Q.; Hu, P.; Murphy, L.C.; Raouf, A. Long Non-Coding RNA H19 Acts as an Estrogen Receptor Modulator that is Required for Endocrine Therapy Resistance in ER+ Breast Cancer Cells. Cell. Physiol. Biochem. 2018, 51, 1518–1532. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Hao, G.; Sun, Y.; Li, L.; Wang, Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. OncoTargets Ther. 2018, 11, 8001–8012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glick, D.; Barth, S.; MacLeod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.-J.; Lei, Y.-H.; Yao, N.; Wang, C.-R.; Hu, N.; Ye, W.-C.; Zhang, D.-M.; Chen, Z.-S. Autophagy and multidrug resistance in cancer. Chin. J. Cancer 2017, 36, 1–10. [Google Scholar] [CrossRef]
- Wilde, L.; Tanson, K.; Curry, J.; Martinez-Outschoorn, U. Autophagy in cancer: A complex relationship. Biochem. J. 2018, 475, 1939–1954. [Google Scholar] [CrossRef]
- Li, X.; Wu, Y.; Liu, A.; Tang, X. Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1α feedback regulatory loop. Tumor Biol. 2016, 37, 14733–14743. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, G.; Yang, L.; Qu, J.; Yang, Z.; Zhou, X. Knockdown of Long Non-Coding RNA UCA1 Increases the Tamoxifen Sensitivity of Breast Cancer Cells through Inhibition of Wnt/β-Catenin Pathway. PLoS ONE 2016, 11, e0168406. [Google Scholar] [CrossRef]
- Wu, C.; Luo, J. Long Non-Coding RNA (lncRNA) Urothelial Carcinoma-Associated 1 (UCA1) Enhances Tamoxifen Resistance in Breast Cancer Cells via Inhibiting mTOR Signaling Pathway. Med. Sci. Monit. 2016, 22, 3860–3867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.G.; Yang, M.F.; Ren, Y.Q.; Wu, C.H.; Wang, L.Q. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4362–4368. [Google Scholar] [PubMed]
- Zhang, Y.; Xu, L.; Li, A.; Han, X. The roles of ZEB1 in tumorigenic progression and epigenetic modifications. Biomed. Pharmacother. 2019, 110, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, D.; Li, H.; Lv, Y.; Li, S. Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int. J. Oncol. 2019, 54, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Cai, Y.; He, J. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. OncoTargets Ther. 2015, 8, 2657–2664. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Li, Y.; Wu, Y.; Wang, Y.; Nian, W. Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF- β signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 706–714. [Google Scholar]
- Cai, Y.; He, J.; Zhang, D. Супрессия экспрессии длиннoй некoдирующей РНК CCAT2 пoвышает oтвет на тамoксифен клетoк рака мoлoчнoй железы, резистентных к тамoксифену. Мoлекулярная биoлoгия 2016, 50, 821–827. [Google Scholar] [CrossRef]
- Sun, W.; Li, A.-Q.; Zhou, P.; Jiang, Y.-Z.; Jin, X.; Liu, Y.-R.; Guo, Y.-J.; Yang, W.-T.; Shao, Z.-M.; Xu, X.-E. DSCAM-AS1 regulates the G1 /S cell cycle transition and is an independent prognostic factor of poor survival in luminal breast cancer patients treated with endocrine therapy. Cancer Med. 2018, 7, 6137–6146. [Google Scholar] [CrossRef] [Green Version]
- Niknafs, Y.S.; Han, S.; Ma, T.; Speers, C.; Zhang, C.; Wilder-Romans, K.; Iyer, M.K.; Pitchiaya, S.; Malik, R.; Hosono, Y.; et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat. Commun. 2016, 7, 12791. [Google Scholar] [CrossRef]
- Ma, Y.; Bu, D.; Long, J.; Chai, W.; Dong, J. LncRNA DSCAM-AS1 acts as a sponge of miR-137 to enhance Tamoxifen resistance in breast cancer. J. Cell. Physiol. 2018, 234, 2880–2894. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yang, Y.A.; Zhang, A.; Fong, K.-W.; Kim, J.; Song, B.; Li, S.; Zhao, J.C.; Yu, J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2015, 35, 2746–2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Kasiri, S.; Bashyal, A.; Mandal, S.S. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J. Mol. Biol. 2013, 425, 3707–3722. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.; Qu, K.; Zhong, F.; Artandi, S.E.; Chang, H.Y. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Zhang, H.-C.; Li, L.; Li, C.-X.; Di, X.; Qu, X. Downregulation of Long Noncoding RNA HOTAIR and EZH2 Induces Apoptosis and Inhibits Proliferation, Invasion, and Migration of Human Breast Cancer Cells. Cancer Biother. Radiopharm. 2018, 33, 241–251. [Google Scholar] [CrossRef]
- Ma, T.; Liang, Y.; Li, Y.; Song, X.; Zhang, N.; Li, X.; Chen, B.; Zhao, W.; Wang, L.; Yang, Q. LncRNA LINP1 confers tamoxifen resistance and negatively regulated by ER signaling in breast cancer. Cell. Signal. 2020, 68, 109536. [Google Scholar] [CrossRef]
- Lin, X.; Dinglin, X.; Cao, S.; Zheng, S.; Wu, C.; Chen, W.; Li, Q.; Hu, Q.; Zheng, F.; Wu, Z.; et al. Enhancer-Driven lncRNA BDNF-AS Induces Endocrine Resistance and Malignant Progression of Breast Cancer through the RNH1/TRIM21/mTOR Cascade. Cell Rep. 2020, 31, 107753. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wen, T.; Li, Z.; Feng, L.; Zhou, L.; Yang, Z.; Xu, L.; Shi, S.; Hou, K.; Shen, J.; et al. Cross-talk between the ER pathway and the lncRNA MAFG-AS1/miR-339-5p/ CDK2 axis promotes progression of ER+ breast cancer and confers tamoxifen resistance. Aging 2020, 12, 20658–20683. [Google Scholar] [CrossRef]
- Shi, Q.; Li, Y.; Li, S.; Jin, L.; Lai, H.; Wu, Y.; Cai, Z.; Zhu, M.; Li, Q.; Li, Y.; et al. LncRNA DILA1 inhibits Cyclin D1 degradation and contributes to tamoxifen resistance in breast cancer. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Li, H.; Zhang, G.Y.; Pan, C.H.; Zhang, X.Y.; Su, X.Y. LncRNA MAFG-ASl promotes the aggressiveness of breast carcinoma through regulating miR-339-5p/MMP. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2838–2846. [Google Scholar] [CrossRef]
- Jia, H.; Wu, D.; Zhang, Z.; Li, S. Regulatory effect of the MAFG-AS1/miR-150-5p/MYB axis on the proliferation and migration of breast cancer cells. Int. J. Oncol. 2020, 58, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Fu, Y.; Guo, F.; Chen, H.; Fu, X.; Tan, W.; Zhang, H. Long non-coding RNA MAFG-AS1 knockdown blocks malignant progression in breast cancer cells by inactivating JAK2/STAT3 signaling pathway via MAFG-AS1/miR-3196/TFAP2A axis. Int. J. Clin. Exp. Pathol. 2020, 13, 2455–2473. [Google Scholar] [PubMed]
- Shi, Y.-F.; Lu, H.; Wang, H.-B. Downregulated lncRNA ADAMTS9-AS2 in breast cancer enhances tamoxifen resistance by activating microRNA-130a-5p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1563–1573. [Google Scholar]
- Gu, J.; Wang, Y.; Wang, X.; Zhou, D.; Shao, C.; Zhou, M.; He, Z. RETRACTED: Downregulation of lncRNA GAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer Lett. 2018, 434, 1–10. [Google Scholar] [CrossRef]
- Lee, M.-S.; Jeong, M.-H.; Lee, H.-W.; Han, H.-J.; Ko, A.; Hewitt, S.; Kim, J.-H.; Chun, K.-H.; Chung, J.-Y.; Lee, C.; et al. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat. Commun. 2015, 6, 7769. [Google Scholar] [CrossRef]
- Xu, C.; Kong, X.; Wang, H.; Zhang, N.; Kong, X.; Ding, X.; Li, X.; Yang, Q. MTDH Mediates Estrogen-Independent Growth and Tamoxifen Resistance by Down-Regulating PTEN in MCF-7 Breast Cancer Cells. Cell. Physiol. Biochem. 2014, 33, 1557–1567. [Google Scholar] [CrossRef]
- Song, E.-L.; Xing, L.; Wang, L.; Song, W.-T.; Li, D.-B.; Wang, Y.; Gu, Y.-W.; Liu, M.-M.; Ni, W.-J.; Zhang, P.; et al. LncRNA ADAMTS9-AS2 inhibits cell proliferation and decreases chemoresistance in clear cell renal cell carcinoma via the miR-27a-3p/FOXO1 axis. Aging 2019, 11, 5705–5725. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Xu, Z.; Chen, X.; Wang, X.; Zeng, S.; Zhao, Z.; Qian, L.; Li, Z.; Wei, J.; Huo, L.; et al. Novel Function of lncRNA ADAMTS9-AS2 in Promoting Temozolomide Resistance in Glioblastoma via Upregulating the FUS/MDM2 Ubiquitination Axis. Front. Cell Dev. Biol. 2019, 7, 217. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, J.; Dong, L.; Ma, R. LncRNA ATXN8OS enhances tamoxifen resistance in breast cancer. Open Med. 2020, 16, 68–80. [Google Scholar] [CrossRef]
- Qu, R.; Hu, C.; Tang, Y.; Yu, Q.; Shi, G. Long Non-coding RNA BLACAT1 Induces Tamoxifen Resistance in Human Breast Cancer by Regulating miR-503/Bcl-2 Axis. Cancer Manag. Res. 2020, 12, 1771–1777. [Google Scholar] [CrossRef] [Green Version]
- Ríos-Romero, M.; Cedro-Tanda, A.; Peña-Luna, M.; Mancera-Rodríguez, M.A.; Hidalgo-Pérez, L.; Cisneros-Villanueva, M.; Beltrán-Anaya, F.O.; Arellano-Llamas, R.; Jiménez-Morales, S.; Alfaro-Ruíz, L.A.; et al. FAM83H-AS1 is a potential modulator of cancer driver genes across different tumors and a prognostic marker for ER/PR + BRCA patients. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Kim, C.Y.; Oh, J.H.; Lee, J.-Y.; Kim, M.H. The LncRNA HOTAIRM1 Promotes Tamoxifen Resistance by Mediating HOXA1 Expression in ER+ Breast Cancer Cells. J. Cancer 2020, 11, 3416–3423. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xu, X.; Jiang, Y.; Jin, X.; Zhou, P.; Liu, Y.; Guo, Y.; Ma, D.; Zuo, W.; Huang, S.; et al. Transcriptome analysis of Luminal Breast Cancer Reveals a Role for LOL in Tumor Progression and Tamoxifen Resistance. Int. J. Cancer 2019, 145, 842–856. [Google Scholar] [CrossRef]
- Mitobe, Y.; Ikeda, K.; Suzuki, T.; Takagi, K.; Kawabata, H.; Horie-Inoue, K.; Inoue, S. ESR1-Stabilizing Long Noncoding RNA TMPO-AS1 Promotes Hormone-Refractory Breast Cancer Progression. Mol. Cell. Biol. 2019, 39. [Google Scholar] [CrossRef]
- Wang, Z.; Katsaros, D.; Biglia, N.; Shen, Y.; Loo, L.; Yu, X.; Lin, H.; Fu, Y.; Chu, W.-M.; Fei, P.; et al. ERα upregulates the expression of long non-coding RNA LINC00472 which suppresses the phosphorylation of NF-κB in breast cancer. Breast Cancer Res. Treat. 2019, 175, 353–368. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, M.; Sun, H.; Zhu, T.; Wang, X. Downregulation of LINC00894-002 Contributes to Tamoxifen Resistance by Enhancing the TGF-β Signaling Pathway. Biochemistry 2018, 83, 603–611. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kääb, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs – Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 1–25. [Google Scholar] [CrossRef]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barazetti, J.F.; Jucoski, T.S.; Carvalho, T.M.; Veiga, R.N.; Kohler, A.F.; Baig, J.; Al Bizri, H.; Gradia, D.F.; Mader, S.; Carvalho de Oliveira, J. From Micro to Long: Non-Coding RNAs in Tamoxifen Resistance of Breast Cancer Cells. Cancers 2021, 13, 3688. https://doi.org/10.3390/cancers13153688
Barazetti JF, Jucoski TS, Carvalho TM, Veiga RN, Kohler AF, Baig J, Al Bizri H, Gradia DF, Mader S, Carvalho de Oliveira J. From Micro to Long: Non-Coding RNAs in Tamoxifen Resistance of Breast Cancer Cells. Cancers. 2021; 13(15):3688. https://doi.org/10.3390/cancers13153688
Chicago/Turabian StyleBarazetti, Jéssica Fernanda, Tayana Shultz Jucoski, Tamyres Mingorance Carvalho, Rafaela Nasser Veiga, Ana Flávia Kohler, Jumanah Baig, Hend Al Bizri, Daniela Fiori Gradia, Sylvie Mader, and Jaqueline Carvalho de Oliveira. 2021. "From Micro to Long: Non-Coding RNAs in Tamoxifen Resistance of Breast Cancer Cells" Cancers 13, no. 15: 3688. https://doi.org/10.3390/cancers13153688
APA StyleBarazetti, J. F., Jucoski, T. S., Carvalho, T. M., Veiga, R. N., Kohler, A. F., Baig, J., Al Bizri, H., Gradia, D. F., Mader, S., & Carvalho de Oliveira, J. (2021). From Micro to Long: Non-Coding RNAs in Tamoxifen Resistance of Breast Cancer Cells. Cancers, 13(15), 3688. https://doi.org/10.3390/cancers13153688