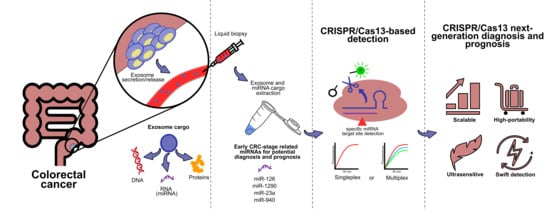
CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. CRC Relevance, Risk Factors, and Key Stages for Diagnostic Survival
3. Current CRC Diagnosis and Their Challenges: Traditional and Molecular Methods
4. Current Clinical Molecular Biomarkers for CRC
5. Extracellular Vesicles as Potential Molecular Biomarkers for Early Diagnosis
6. CEx-miRNAs for CRC Diagnosis
7. CRISPR/Cas Systems
8. CRISPR/Cas13-Based Platforms as a Potential Candidate for CRC Early Diagnosis and Prognosis
9. Dedicated crRNA Design for a Potential CRISPR/Cas13-Based Platform for CRC miRNAs-Based Diagnosis
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lauby-Secretan, B.; Vilahur, N.; Bianchini, F.; Guha, N.; Straif, K.; International Agency for Research on Cancer Handbook Working Group. The IARC perspective on colorectal cancer screening. N. Engl. J. Med. 2018, 378, 1734–1740. [Google Scholar] [CrossRef] [PubMed]
- Barani, M.; Bilal, M.; Rahdar, A.; Arshad, R.; Kumar, A.; Hamishekar, H.; Kyzas, G.Z. Nanodiagnosis and nanotreatment of colorectal cancer: An overview. J. Nanopart. Res. 2021, 23, 18. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Angell, H.K.; Bruni, D.; Barrett, J.C.; Herbst, R.; Galon, J. The immunoscore: Colon cancer and beyond. Clin. Cancer Res. 2020, 26, 332–339. [Google Scholar] [CrossRef] [Green Version]
- Zarour, L.R.; Anand, S.; Billingsley, K.G.; Bisson, W.H.; Cercek, A.; Clarke, M.F.; Coussens, L.M.; Gast, C.E.; Geltzeiler, C.B.; Hansen, L.; et al. Colorectal Cancer Liver Metastasis: Evolving Paradigms and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 163–173. [Google Scholar] [CrossRef] [Green Version]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral bacteria and intestinal dysbiosis in colorectal cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef] [Green Version]
- Lew, J.-B.; Feletto, E.; Wade, S.; Caruana, M.; Kang, Y.-J.; Nickson, J.; Simms, K.T.; Procopio, P.; Taylor, N.; Worthington, J.; et al. Benefits, harms and cost-effectiveness of cancer screening in Australia: An overview of modelling es-timates. Public Health Res. Pract. 2019, 29, 29121913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.-H.; Chen, Y.-X.; Fang, J.-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target Ther. 2020, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Gungormez, C.; Aktas, H.; Dilsiz, N.; Borazan, E. Novel miRNAs as potential biomarkers in stage II colon cancer: Microarray analysis. Mol. Biol. Rep. 2019, 46, 4175–4183. [Google Scholar] [CrossRef]
- Chen, W.-Y.; Zhao, X.-J.; Yu, Z.-F.; Hu, F.-L.; Liu, Y.-P.; Cui, B.-B.; Dong, X.-S.; Zhao, Y.S. The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 7092–7101. [Google Scholar]
- Hibner, G.; Kimsa-Furdzik, M.; Francuz, T. Relevance of MicroRNAs as potential diagnostic and prognostic markers in colo-rectal cancer. Int. J. Mol. Sci. 2018, 19, 2944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, J.V.; Galbraith, N.J.; Yang, D.; Burton, J.F.; Walker, S.P.; Galandiuk, S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: A systematic review and meta-analysis. Br. J. Cancer 2017, 116, 762–774. [Google Scholar] [CrossRef]
- Desmond, B.J.; Dennett, E.R.; Danielson, K.M. Circulating extracellular vesicle microRNA as diagnostic biomarkers in early colorectal cancer—A review. Cancers 2020, 12, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, L.; Zhu, S.; Chen, L.; Liu, X.; Wei, R.; Zhao, L.; Yang, Y.; Zhang, Z.; Kong, G.; Li, P.; et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: A comparison with plasma total miRNAs. J. Extracell. Vesicles 2019, 8, 1643670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Zhong, J.; Zhong, B.; Huang, J.; Jiang, L.; Jiang, Y.; Yuan, J.; Sun, J.; Dai, L.; Yang, C.; et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 476, 13–22. [Google Scholar] [CrossRef]
- Liu, W.; Yang, D.; Chen, L.; Liu, Q.; Wang, W.; Yang, Z.; Shang, A.; Quan, W.; Li, D. Plasma Exosomal miRNA-139-3p is a Novel Biomarker of Colorectal Cancer. J. Cancer 2020, 11, 4899–4906. [Google Scholar] [CrossRef]
- Francavilla, A.; Turoczi, S.; Tarallo, S.; Vodicka, P.; Pardini, B.; Naccarati, A. Exosomal microRNAs and other non-coding RNAs as colorectal cancer biomarkers: A review. Mutagenesis 2020, 35, 243–260. [Google Scholar] [CrossRef]
- Egloff, S.; Melnychuk, N.; Reisch, A.; Martin, S.; Klymchenko, A.S. Enzyme-free amplified detection of cellular microRNA by light-harvesting fluorescent nanoparticle probes. Biosens. Bioelectron. 2021, 179, 113084. [Google Scholar] [CrossRef]
- Pang, S.-W.; Awi, N.J.; Armon, S.; Lim, W.W.-D.; Low, J.S.-H.; Peh, K.-B.; Peh, S.-C.; Teow, S.-Y. Current update of laboratory molecular diagnostics advancement in management of colorectal cancer (CRC). Diagnostics 2020, 10, 9. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Ezzletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 241–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chertow, D.S. CRISPR Portable Diagnostic Tools. Science 2018, 360, 381–383. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Prot. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [Green Version]
- Bruch, R.; Urban, G.A.; Dincer, C. CRISPR/Cas Powered Multiplexed Biosensing. Trends Biotechnol. 2019, 37, 791–792. [Google Scholar] [CrossRef]
- Zuo, X.; Fan, C.; Chen, H.Y. Biosensing: CRISPR-powered diagnostics. Nat. Biomed. Eng. 2017, 1, 0091. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef]
- Burmistrz, M.; Krakowski, K.; Krawczyk-Balska, A. RNA-targeting CRISPR–Cas systems and their applications. Int. J. Mol. 2020, 21, 1122. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Mukama, O.; Wu, J.; Li, Z.; Liang, Q.; Yi, Z.; Lu, X.; Liu, Y.; Liu, Y.; Hussain, M.; Makafe, G.G.; et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020, 10, 112143. [Google Scholar] [CrossRef]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Brenner, H.; Chen, C. The colorectal cancer epidemic: Challenges and opportunities for primary, secondary and tertiary pre-vention. Br. J. Cancer 2018, 119, 785–792. [Google Scholar] [CrossRef] [Green Version]
- International Agency for Research on Cancer (IARC). Globocan 2020. Available online: https://gco.iarc.fr/today/home (accessed on 14 July 2021).
- Nguyen, H.T.; Duong, H.Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy (review). Oncol. Lett. 2018, 16, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Czene, K.; Lichtenstein, P.; Hemminki, K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int. J. Cancer 2002, 99, 260–266. [Google Scholar] [CrossRef]
- Lichtenstein, P.; Holm, N.V.; Verkasalo, P.K.; Iliadou, A.; Kaprio, J.; Koskenvou, M.; Pukkala, E.; Skytthe, A.; Hermminki, K. Environmental and heritable factors in the causation of cancer—Analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000, 343, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Keum, N.N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef] [PubMed]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153–173. [Google Scholar] [PubMed]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing; American Joint Commission on Cancer: Chicago, IL, USA, 2017; pp. 251–274. [Google Scholar]
- National Cancer Institute: Diagnosis and Staging. Available online: https://www.cancer.gov/about-cancer/diagnosis-staging/staging (accessed on 6 September 2021).
- National Cancer Institute: Surveillance, Epidemiology, and End Results Program (SEER 2017)—Statistics At a Glance. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 9 July 2021).
- Idos, G.E.; Kwok, J.; Bonthala, N.; Kysh, L.; Gruber, S.B.; Qu, C. The Prognostic Implications of Tumor Infiltrating Lymphocytes in Colorectal Cancer: A Systematic Review and Meta-Analysis. Sci. Rep. 2020, 10, 3360. [Google Scholar] [CrossRef]
- Das, V.; Kalita, J.; Pal, M. Predictive and prognostic biomarkers in colorectal cancer: A systematic review of recent advances and challenges. Biomed. Pharmacother. 2017, 87, 8–19. [Google Scholar] [CrossRef]
- Boakye, D.; Rillmann, B.; Walter, V.; Jansen, L.; Hoffmeister, M.; Brenner, H. Impact of comorbidity and frailty on prognosis in colorectal cancer patients: A systematic review and meta-analysis. Cancer Treat. Rev. 2018, 64, 30–39. [Google Scholar] [CrossRef]
- White, A.; Ironmonger, L.; Steele, R.J.C.; Ormiston-Smith, N.; Crawford, C.; Seims, A. A review of sex-related differences in colorectal cancer incidence, screening uptake, routes to diagnosis, cancer stage and survival in the UK. BMC Cancer 2018, 18, 906. [Google Scholar] [CrossRef] [Green Version]
- Brouwer, N.P.M.; Bos, A.C.R.K.; Lemmens, V.E.P.P.; Tanis, P.J.; Hugen, N.; Nagtegaal, I.D.; de Wilt, J.H.W.; Verhoeven, R.H.A. An overview of 25 years of incidence, treatment and outcome of colorectal cancer patients. Int. J. Cancer 2018, 143, 2758–2766. [Google Scholar] [CrossRef]
- Marcuello, M.; Vymetalkova, V.; Neves, R.P.L.; Duran-Sanchon, S.; Vedeld, H.M.; Tham, E.; van Dalum, G.; Flügen, G.; Garcia-Barberan, V.; Fijneman, R.J.; et al. Circu-lating biomarkers for early detection and clinical management of colorectal cancer. Mol. Asp. Med. 2019, 69, 107–122. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Socierty, Colorectal Cancer–Early Detection, Diagnosis and Staging. Available online: https://www.cancer.org/cancer/colon-rectal-cancer/detection-diagnosis-staging/detection.html (accessed on 14 July 2021).
- Wang, Y.W.; Chen, H.H.; Wu, M.S.; Chiu, H.M. Current status and future challenge of population-based organized colorectal cancer screening: Lesson from the first decade of Taiwanese program. J. Formos. Med. Assoc. 2018, 117, 358–364. [Google Scholar] [CrossRef]
- Robertson, D.J.; Ladabaum, U. Opportunities and Challenges in Moving from Current Guidelines to Personalized Colorectal Cancer Screening. Gastroenterology 2019, 156, 904–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, X.; Wang, T.; Chen, Z.Y.; Zhang, K.H. Blood-derived molecular signatures as biomarker panels for the early detection of colorectal cancer. Mol. Biol. Rep. 2020, 47, 8159–8168. [Google Scholar] [CrossRef]
- Lurvink, R.J.; Tajzai, R.; Rovers, K.P.; Wassenaar, E.C.E.; Moes, D.J.A.R.; Pluimakers, G.; Boerma, D.; Burger, J.W.A.; Nienhuijs, S.W.; de Hingh, I.H.J.T.; et al. Systemic Pharmacokinetics of Oxaliplatin After Intraperitoneal Admin-istration by Electrostatic Pressurized Intraperitoneal Aerosol Chemotherapy (ePIPAC) in Patients with Unresectable Colorectal Peritoneal Metastases in the CRC-PIPAC Trial. Ann. Surg. Oncol. 2021, 28, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Petrelli, F.; Comito, T.; Barni, S.; Pancera, G.; Scorsetti, M.; Ghidini, A. Stereotactic body radiotherapy for colorectal cancer liver metastases: A systematic review. Radiother. Oncol. 2018, 129, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Oliveres, H.; Pesántez, D.; Maurel, J. Lessons to learn for adequate targeted therapy development in metastatic colorectal cancer patients. Int. J. Mol. Sci. 2021, 22, 5019. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Saraf, S.; Jain, A.; Panda, P.K.; Verma, A.; Jain, S.K. Basics to advances in nanotherapy of colorectal cancer. Drug Deliv. Transl. Res. 2020, 10, 319–338. [Google Scholar] [CrossRef]
- Molinari, C.; Marisi, G.; Passardi, A.; Matteucci, L.; De Maio, G.; Ulivi, P. Heterogeneity in colorectal cancer: A challenge for personalized medicine? Int. J. Mol. Sci. 2018, 19, 3733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahasneh, A.; Al-Shaheri, F.; Jamal, E. Molecular biomarkers for an early diagnosis, effective treatment and prognosis of colorectal cancer: Current updates. Exp. Mol. Pathol. 2017, 102, 475–483. [Google Scholar] [CrossRef]
- Loktionov, A. Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins. World J. Gastrointest. Oncol. 2020, 12, 124–148. [Google Scholar] [CrossRef] [PubMed]
- Moghimi-Dehkordi, B.; Safaee, A. An overview of colorectal cancer survival rates and prognosis in Asia. World J. Gastrointest. Oncol. 2020, 4, 71–75. [Google Scholar] [CrossRef]
- Simon, K. Colorectal cancer development and advances in screening. Clin. Interv. Aging 2016, 11, 967–976. [Google Scholar]
- Nikolouzakis, T.K.; Vassilopoulou, L.; Fragkiadaki, P.; Sapsakos, T.M.; Papadakis, G.Z.; Spandidos, D.A.; Tsatsakis, A.M.; Tsiaoussis, J. Improving diagnosis, prognosis and prediction by using biomarkers in CRC patients (Review). Oncol. Rep. 2018, 39, 2455–2472. [Google Scholar] [CrossRef] [Green Version]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Martini, G.; Dienstmann, R.; Ros, J.; Baraibar, I.; Cuadra-Urteaga, J.L.; Salva, F.; Ciardiello, D.; Mulet, N.; Argiles, G.; Tab-ernero, J.; et al. Molecular subtypes and the evolution of treatment management in metastatic colorectal cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920936089. [Google Scholar] [CrossRef]
- Amaro, A.; Chiara, S.; Pfeffer, U. Molecular Evolution of Colorectal Cancer: From Multistep Carcinogenesis to the Big Bang. Cancer Metastasis Rev. 2016, 35, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; O’Brien, C.A.; van Galen, P.; Gan, O.I.; Notta, F.; Brown, A.M.; Ng, K.; Ma, J.; Wienholds, E.; Dunant, C.; et al. Variable Clonal Repopulation Dynamics Influence Chemotherapy Response in Colorectal Cancer. Science 2013, 339, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stintzing, S.; Tejpar, S.; Gibbs, P.; Thiebach, L.; Lenz, H.J. Understanding the Role of Primary Tumour Localisation in Colorectal Cancer Treatment and Outcomes. Eur. J. Cancer 2017, 84, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Z.; Yu, X.; Yang, B.; Zhang, Y.; Zhang, L.; Li, X.; Sun, H. Colorectal cancer heterogeneity and targeted therapy: Clinical implications, challenges and solutions for treatment resistance. Semin. Cell Dev. Biol. 2017, 64, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Issa, I.A.; NouredDine, M. Colorectal cancer screening: An updated review of the available options. World J. Gastrointest. Oncol. 2017, 23, 5086–5096. [Google Scholar] [CrossRef] [PubMed]
- Kavousipour, S.; Khademi, F.; Zamani, M.; Vakili, B.; Mokarram, P. Novel biotechnology approaches in colorectal cancer diagnosis and therapy. Biotechnol. Lett. 2017, 39, 785–803. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; De Brabander, I.; Francart, J.; Candeur, M.; Polus, M.; Van Eycken, L.; Brenner, H. Benefits of switching from guai-ac-based faecal occult blood to faecal immunochemical testing: Experience from the Wallonia–Brussels colorectal cancer screening programme. Br. J. Cancer 2020, 122, 1109–1117. [Google Scholar] [CrossRef]
- Carethers, J.M. Fecal DNA Testing for Colorectal Cancer Screening. Annu. Rev. Med. 2020, 71, 59–69. [Google Scholar] [CrossRef]
- Rex, D.K.; Boland, C.R.; Dominitz, J.A.; Giardiello, F.M.; Johnson, D.A.; Kaltenbach, T.; Levin, T.R.; Lieberman, D.; Robertson, D.J. Colorectal Cancer Screening: Recommendations for Physicians and Patients from the U.S. Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2017, 153, 307–323. [Google Scholar] [CrossRef]
- Robbins, E.C.; Cross, A.J. Guaiac Fecal Occult Blood Tests and Mortality: A 30-Year Follow-up of Two Pooled Trials. Clin. Gastroenterol. Hepatol. 2021, 19, 892–894. [Google Scholar] [CrossRef]
- Pickhardt, P.J.; Graffy, P.M.; Weigman, B.; Deiss-Yehiely, N.; Hassan, C.; Weiss, J.M. Diagnostic performance of multitarget stool DNA and CT colonography for noninvasive colorectal cancer screening. Radiology 2020, 297, 120–129. [Google Scholar] [CrossRef]
- Lin, Z.; Chen, Y.; Lin, Y.; Lin, H.; Li, H.; Su, X.; Fang, Z.; Wang, J.; Wei, Q.; Teng, J.; et al. Potential miRNA biomarkers for the diagnosis and prognosis of esophageal cancer detected by a novel absolute quantitative RT-qPCR method. Sci. Rep. 2020, 10, 20065. [Google Scholar] [CrossRef]
- Ilie, M.; Butori, C.; Lassalle, S.; Heeke, S.; Piton, N.; Sabourin, J.C.; Tanga, V.; Washetine, K.; Long-Mira, E.; Maitre, P.; et al. Optimization of EGFR mutation detection by the fully-automated qPCR-based Idylla system on tumor tissue from patients with non-small cell lung cancer. Oncotarget 2017, 8, 103055–103062. [Google Scholar] [CrossRef] [Green Version]
- Rochow, H.; Franz, A.; Jung, M.; Weickmann, S.; Ralla, B.; Kilic, E.; Stephan, C.; Fendler, A.; Jung, K. Instability of circular RNAs in clinical tissue samples impairs their reliable expression analysis using RT-qPCR: From the myth of their advantage as biomarkers to reality. Theranostics 2020, 10, 9268–9279. [Google Scholar] [CrossRef] [PubMed]
- Vanova, B.; Kalman, M.; Jasek, K.; Kasubova, I.; Burjanivova, T.; Farkasova, A.; Kruzliak, P.; Busselberg, D.; Plank, L.; Lasabova, Z. Droplet digital PCR revealed high concordance between primary tumors and lymph node metastases in multiplex screening of KRAS mutations in colorectal cancer. Clin. Exp. Med. 2019, 19, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Tavano, F.; Gioffreda, D.; Valvano, M.R.; Palmieri, O.; Tardio, M.; Latiano, T.P.; Piepoli, A.; Maiello, E.; Pirozzi, F.; Andriulli, A. Droplet digital PCR quantification of miR-1290 as a circulating biomarker for pancreatic cancer. Sci. Rep. 2018, 8, 16389. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, S.; Damin, F.; Ferraro, L.; Soriani, N.; Burgio, V.; Ronzoni, M.; Gianni, L.; Ferrari, M.; Chiari, M. Microarray Approach Combined with ddPCR: An Useful Pipeline for the Detection and Quantification of Circulating Tumour dna Mutations. Cells 2019, 8, 769. [Google Scholar] [CrossRef] [Green Version]
- Del Vecchio, F.; Mastroiaco, V.; Di Marco, A.; Compagnoni, C.; Capece, D.; Zazzeroni, F.; Capalbo, C.; Alesse, E.; Tessitore, A. Next-generation sequencing: Recent applications to the analysis of colorectal cancer. J. Transl. Med. 2017, 15, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Zhang, R.; Li, J. CRISPR/cas systems redefine nucleic acid detection: Principles and methods. Biosens. Bioelectron. 2020, 165, 112430. [Google Scholar] [CrossRef]
- Cheng, Y.; Dong, L.; Zhang, J.; Zhao, Y.; Li, Z. Recent advances in microRNA detection. Analyst 2018, 143, 1758–1774. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Fan, T.; Chen, Y.; Ye, L.; Zhang, C.; Liu, F.; Qin, Y.; Tan, Y.; Jiang, Y. Zeptomolar-level one-pot simultaneous detection of multiple colorectal cancer microRNAs by cascade isothermal amplification. Biosens. Bioelectron. 2020, 169, 112631. [Google Scholar] [CrossRef]
- Bonini, A.; Poma, N.; Vivaldi, F.; Kirchhain, A.; Salvo, P.; Bottai, D.; Tavanti, A.; Di Francesco, F. Advances in biosensing: The CRISPR/Cas system as a new powerful tool for the detection of nucleic acids. J. Pharm. Biomed. Anal. 2021, 192, 113645. [Google Scholar] [CrossRef]
- Latacz, M.; Snarska, J.; Kostyra, E.; Wroński, K.; Fiedorowicz, E.; Savelkoul, H.; Jarmołowska, B.; Płomiński, J.; Grzybowski, R.; Cieślińska, A. CYP27B1 Gene Polymorphism rs10877012 in Patients Diagnosed with Colorectal Cancer. Nutrients 2020, 12, 998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwaenepoel, K.; Holmgaard Duelund, J.; De Winne, K.; Maes, V.; Weyn, C.; Lambin, S.; Dendooven, R.; Broeckx, G.; Steiniche, T.; Pauwels, P. Clinical Performance of the Idylla MSI Test for a Rapid Assessment of the DNA Microsatellite Status in Human Colorectal Cancer. J. Mol. Diagn. 2020, 22, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.X.; Fu, Q.; Guo, Y.Y.; Ye, M.; Zhao, H.X.; Wang, Q.; Peng, X.M.; Li, Q.W.; Wang, R.L.; Xiao, W.H. Effectiveness of circulating tumor DNA for detection of KRAS gene mutations in colorectal cancer patients: A meta-analysis. OncoTargets Ther. 2017, 10, 945–953. [Google Scholar] [CrossRef] [Green Version]
- Loupakis, F.; Moretto, R.; Aprile, G.; Muntoni, M.; Cremolini, C.; Iacono, D.; Casagrande, M.; Ferrari, L.; Salvatore, L.; Schirripa, M.; et al. Clinico-pathological nomogram for predicting BRAF mutational status of metastatic colorectal cancer. Br. J. Cancer 2016, 114, 30–36. [Google Scholar] [CrossRef]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular biomarkers for the evaluation of colorectal cancer: Guideline from The American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. Am. J. Clin. Oncol. 2017, 35, 1453–1496. [Google Scholar] [CrossRef]
- Ho, H.-H.; Joo, Y.-E. Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intest. Res. 2019, 18, 168–183. [Google Scholar]
- Cha, B.S.; Park, K.S.; Park, J.S. Signature mRNA markers in extracellular vesicles for the accurate diagnosis of colorectal cancer. J. Biol. Eng. 2020, 14, 4. [Google Scholar] [CrossRef]
- Kim, M.; Lee, S.T.; Choi, S.; Lee, H.; Kwon, S.S.; Byun, J.H.; Kim, Y.A.; Rhee, K.J.; Choi, J.R.; Kim, T.I.; et al. Fusobacterium nucleatum in biopsied tissues from colorectal cancer patients and alcohol consumption in Korea. Sci. Rep. 2020, 10, 19915. [Google Scholar] [CrossRef]
- Shang, A.; Gu, C.; Wang, W.; Wang, X.; Sun, J.; Zeng, B.; Chen, C.; Chang, W.; Ping, Y.; Ji, P.; et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol. Cancer 2020, 19, 117. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on extracellular vesicles: Introducing the next small big thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalishwaralal, K.; Kwon, W.Y.; Park, K.S. Exosomes for non-invasive Cancer monitoring. Biotechnol. J. 2019, 14, 1800430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Li, Y.; Yuan, Y.; Liu, B.; Pan, S.; Liu, Q.; Qi, X.; Zhou, H.; Dong, W.; Jie, L. The potential of exosomes derived from colorectal cancer as a biomarker. Clin. Chim. Acta 2019, 490, 186–193. [Google Scholar] [CrossRef]
- Koncina, E.; Haan, S.; Rauh, S.; Letellier, E. Prognostic and predictive molecular biomarkers for colorectal cancer: Updates and challenges. Cancers 2020, 12, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacante, M.; Borzì, A.M.; Basile, F.; Biondi, A. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J. Clin. Cases 2019, 6, 869–881. [Google Scholar] [CrossRef]
- Hu, T.; Wolfram, J.; Srivastava, S. Extracellular Vesicles in Cancer Detection: Hopes and Hypes. Trends Cancer 2021, 7, 122–133. [Google Scholar] [CrossRef]
- Vader, P.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Emerging targets for cancer therapy. Trends Mol. Med. 2014, 20, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Xavier, C.P.R.; Caires, H.R.; Barbosa, M.A.G.; Bergantim, R.; Guimarães, J.E.; Vasconcelos, M.H. The Role of Extracellular Vesicles in the Hallmarks of Cancer and Drug Resistance. Cells 2020, 9, 1141. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, M.H.; Caires, H.R.; Abols, A.; Xavier, C.P.R.; Line, A. Extracellular vesicles as a novel source of biomarkers in liquid biopsies for monitoring cancer progression and drug resistance. Drug Resist. Updat. 2019, 47, 100647. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, H.; Harris, A.L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018, 27, 281–298. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, D.; Jin, F.; Bian, Z.; Li, L.; Liang, H.; Li, M.; Shi, L.; Pan, C.; Zhu, D.; et al. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat. Commun. 2017, 8, 14041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Q.; Zhou, L.; Sui, H.; Yang, L.; Wu, X.; Song, Q.; Jia, R.; Li, R.; Sun, J.; Wang, Z.; et al. Primary tumors release ITGBL1-rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast-niche formation. Nat. Commun. 2020, 11, 1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palazzolo, S.; Memeo, L.; Hadla, M.; Duzagac, F.; Steffan, A.; Perin, T.; Canzonieri, V.; Tuccinardi, T.; Caligiuri, I.; Rizzolio, F. Cancer extracellular vesicles: Next-generation diagnostic and drug delivery nanotools. Cancers 2020, 12, 3165. [Google Scholar] [CrossRef] [PubMed]
- Bracci, L.; Lozupone, F.; Parolini, I. The role of exosomes in colorectal cancer disease progression and response to therapy. Cytokine Growth Factor Rev. 2020, 51, 84–91. [Google Scholar] [CrossRef]
- Bahrami, A.; Moradi Binabaj, M.; Ferns, G.A. Exosomes: Emerging modulators of signal transduction in colorectal cancer from molecular understanding to clinical application. Biomed. Pharmacother. 2021, 141, 111882. [Google Scholar] [CrossRef]
- Cheshomi, H.; Matin, M.M. Exosomes and their importance in metastasis, diagnosis, and therapy of colorectal cancer. J. Cell. Biochem. 2019, 120, 2671–2686. [Google Scholar] [CrossRef] [PubMed]
- Vafaei, S.; Roudi, R.; Madjd, Z.; Aref, A.R.; Ebrahimi, M. Potential theranostics of circulating tumor cells and tumor-derived exosomes application in colorectal cancer. Cancer Cell Int. 2020, 20, 288. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bebelman, M.P.; Janssen, E.; Pegtel, D.M.; Crudden, C. The forces driving cancer extracellular vesicle secretion. Neoplasia 2021, 23, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Kroeger, B.; Marie, P.P.; Bridges, E.M.; Mason, J.D.; McCormick, K.; Zois, C.E.; Sheldon, H.; Khalid Alham, N.; Johnson, E.; et al. Glutamine deprivation alters the origin and function of cancer cell exosomes. EMBO J. 2020, 39, e103009. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Acuña, R.A.; Varas-Godoy, M.; Retamal, M.A. Connexin-46 Contained in Extracellular Vesicles Enhance Malignance Char-acteristics in Breast Cancer Cells. Biomolecules 2020, 10, 676. [Google Scholar] [CrossRef]
- Mostafazadeh, M.; Samadi, N.; Kahroba, H.; Baradaran, B.; Haiaty, S.; Nouri, M. Potential roles and prognostic significance of exosomes in cancer drug resistance. Cell Biosci. 2021, 11, 1. [Google Scholar] [CrossRef]
- Drula, R.; Ott, L.F.; Berindan-Neagoe, I.; Pantel, K.; Calin, G.A. Micrornas from liquid biopsy derived extracellular vesicles: Recent advances in detection and characterization methods. Cancers 2020, 12, 2009. [Google Scholar] [CrossRef]
- Han, W.; Cui, H.; Liang, J.; Su, X. Role of MicroRNA-30c in cancer progression. J. Cancer 2020, 11, 2593–2601. [Google Scholar] [CrossRef] [Green Version]
- Noguchi, T.; Toiyama, Y.; Kitajima, T.; Imaoka, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Toden, S.; et al. MiRNA-503 Promotes Tumor Progression and Is Associated with Early Recurrence and Poor Prognosis in Human Colorectal Cancer. Oncology 2016, 90, 221–231. [Google Scholar] [CrossRef]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhang, Z.; Gao, Q. Transfer of microRNA-25 by colorectal cancer cell-derived extracellular vesicles facilitates colo-rectal cancer development and metastasis. Mol. Ther. Nucleic Acids 2021, 23, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhang, Y.; Li, B.; Kang, M.; Yang, Z.; Lin, C.; Hu, K.; Wei, Z.; Xu, M.; Mi, J.; et al. miRNAs derived from circulating small extracellular vesicles as diagnostic biomarkers for nasopharyngeal carcinoma. Cancer Sci. 2021, 112, 2393–2404. [Google Scholar] [CrossRef]
- Gallardo-Gómez, M.; Álvarez-Chaver, P.; Cubiella, J. Colorectal cancer screening and diagnosis: Omics-based technologies for development of a non-invasive blood-based method. Expert Rev. Anticancer Ther. 2021, 21, 723–738. [Google Scholar] [CrossRef]
- Danese, E.; Minicozzi, A.M.; Benati, M.; Paviati, E.; Lima-Oliveira, G.; Gusella, M.; Pasini, F.; Salvagno, G.L.; Montagnana, M.; Lippi, G. Reference miRNAs for colorectal cancer: Analysis and verification of current data. Sci. Rep. 2017, 7, 8413. [Google Scholar] [CrossRef]
- Nassar, F.J.; Msheik, Z.S.; Itani, M.M.; Helou, R.E.; Hadla, R.; Kreidieh, F.; Bejjany, R.; Mukherji, D.; Shamseddine, A.; Nasr, R.R.; et al. Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metastasis. Diagnostics 2021, 11, 341. [Google Scholar] [CrossRef]
- Yan, S.; Han, B.; Gao, S.; Wang, X.; Wang, Z.; Wang, F.; Zhang, J.; Xu, D.; Sun, B. Exosome-encapsulated microRNAs as circulating biomarkers for colorectal cancer. Oncotarget 2017, 8, 60149–60158. [Google Scholar] [CrossRef] [Green Version]
- de Miguel Pérez, D.; Rodriguez Martínez, A.; Ortigosa Palomo, A.; Delgado Ureña, M.; Garcia Puche, J.L.; Robles Remacho, A.; Exposito Hernandez, J.; Lorente Acosta, J.A.; Ortega Sánchez, F.G.; Serrano, M.J. Extracellular vesicle-miRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci. Rep. 2020, 10, 3974. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Zhang, P.P.; Huang, J.J.; Wang, Z.Y.; Zhang, Z.H.; Yuan, J.Z.; Ma, E.M.; Liu, X.; Bai, J. Reduced serum exosomal miR-874 expression predicts poor prognosis in colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 664–672. [Google Scholar]
- Köberle, V.; Pleli, T.; Schmithals, C.; Augusto Alonso, E.; Haupenthal, J.; Bönig, H.; Peveling-Oberhag, J.; Biondi, R.M.; Zeuzem, S.; Kronenberger, B.; et al. Differential Stability of Cell-Free Circulating microRNAs: Implications for Their Utilization as Biomarkers. PLoS ONE 2013, 8, e75184. [Google Scholar] [CrossRef] [Green Version]
- Rahbarghazi, R.; Jabbari, N.; Sani, N.A.; Asghari, R.; Salimi, L.; Kalashani, S.A.; Feghhi, M.; Etemadi, T.; Akbariazar, E.; Mahmoudi, M.; et al. Tumor-derived extracellular vesicles: Reliable tools for Cancer diagnosis and clinical applications. Cell Commun. Signal. 2019, 17, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Zhuang, Y.; Zhang, J.; Chen, M.; Wu, S. Four circulating exosomal miRNAs as novel potential biomarkers for the early diagnosis of human colorectal cancer. Tissue Cell 2021, 70, 101499. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Liu, Y.; Zhang, J.; Bian, Z.; Yao, S.; Fei, B.; Zhou, L.; Yin, Y.; Huang, Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 2019, 84, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Alves dos Santos, K.; Clemente dos Santos, I.C.; Santos Silva, C.; Gomes Ribeiro, H.; de Farias Domingos, I.; Nogueira Silbiger, V. Circulating Exosomal miRNAs as Biomarkers for the Diagnosis and Prognosis of Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 346. [Google Scholar] [CrossRef]
- Zanutto, S.; Ciniselli, C.M.; Belfiore, A.; Lecchi, M.; Masci, E.; Delconte, G.; Primignani, M.; Tosetti, G.; Dal Fante, M.; Fazzini, L.; et al. Plasma miRNA-based signatures in CRC screening programs. Int. J. Cancer 2020, 146, 1164–1173. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; Chen, Y.; Xu, D. Circulating miR-449a predicts survival outcome for colorectal cancer following curative resection: An observational study. Medicine 2021, 100, e25022. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Miao, L. Serum microrna-592 serves as a novel potential biomarker for early diagnosis of colorectal cancer. Oncol. Lett. 2020, 20, 1119–1126. [Google Scholar] [CrossRef]
- Cui, X.; Lv, Z.; Ding, H.; Xing, C.; Yuan, Y. MiR-1539 and Its Potential Role as a Novel Biomarker for Colorectal Cancer. Front. Oncol. 2021, 10, 3449. [Google Scholar] [CrossRef]
- Maminezhad, H.; Ghanadian, S.; Pakravan, K.; Razmara, E.; Rouhollah, F.; Mossahebi-Mohammadi, M.; Babashah, S. A panel of six-circulating miRNA signature in serum and its potential diagnostic value in colorectal cancer. Life Sci. 2020, 258, 118226. [Google Scholar] [CrossRef]
- Wang, N.; Liu, W. Increased expression of miR-552 acts as a potential predictor biomarker for poor prognosis of colorectal cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 412–416. [Google Scholar]
- Fiala, O.; Pitule, P.; Hosek, P.; Liska, V.; Sorejs, O.; Bruha, J.; Vycital, O.; Buchler, T.; Poprach, A.; Topolcan, O.; et al. The association of miR-126-3p, miR-126-5p and miR-664-3p expression profiles with outcomes of patients with metastatic colorectal cancer treated with bevacizumab. Tumor Biol. 2017, 39, 1010428317709283. [Google Scholar] [CrossRef] [Green Version]
- Ulivi, P.; Canale, M.; Passardi, A.; Marisi, G.; Valgiusti, M.; Frassineti, G.L.; Calistri, D.; Amadori, D.; Scarpi, E. Circulating plasma levels of miR-20b, miR-29b and mir-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal cancer. Int. J. Mol. Sci. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, F.; Jiang, W.; Zhou, L.; Chen, Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl. Oncol. 2018, 11, 221–232. [Google Scholar] [CrossRef]
- Tang, Y.; Zhao, Y.; Song, X.; Song, X.; Niu, L.; Xie, L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J. Clin. Lab. Anal. 2019, 33, e23004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- miRBase: The microRNA Database. Available online: http://www.mirbase.org/ (accessed on 18 May 2021).
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, 155–162. [Google Scholar] [CrossRef]
- Gasparello, J.; Papi, C.; Allegretti, M.; Giordani, E.; Carboni, F.; Zazza, S.; Pescarmona, E.; Romania, P.; Giacomini, P.; Scapoli, C.; et al. A distinctive microrna (miRNA) signature in the blood of colorectal cancer (CRC) patients at sur-gery. Cancers 2020, 12, 2410. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, A.; Neefs, I.; Hoeck, S.; Peeters, M.; Van Hal, G. Towards Novel Non-Invasive Colorectal Cancer Screening Methods: A Comprehensive Review. Cancers 2021, 13, 1820. [Google Scholar] [CrossRef]
- Pinzani, P.; D’Argenio, V.; Del Re, M.; Pellegrini, C.; Cucchiara, F.; Salvianti, F.; Galbiati, S. Updates on liquid biopsy: Current trends and future perspectives for clinical application in solid tumors. Clin. Chem. Lab. Med. 2021, 59, 1181–1200. [Google Scholar] [CrossRef]
- Sazanov, A.A.; Kiselyova, E.V.; Zakharenko, A.A.; Romanov, M.N.; Zaraysky, M.I. Plasma and saliva miR-21 expression in colorectal cancer patients. J. Appl. Genet. 2017, 58, 231–237. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Majem, B.; Álvarez-Castro, A.; Díaz-Peña, R.; Abalo, A.; Suárez-Cabrera, L.; Gil-Moreno, A.; Santa-maría, A.; López-López, R.; Muinelo-Romay, L.; et al. A Novel Saliva-Based miRNA Signature for Colo-rectal Cancer Diagnosis. J. Clin. Med. 2019, 8, 2029. [Google Scholar] [CrossRef] [Green Version]
- Burstein, D.; Harrington, L.B.; Strutt, S.C.; Probst, A.J.; Anantharaman, K.; Thomas, B.C.; Doudna, J.A.; Banfield, J.F. New CRISPR-Cas systems from uncultivated microbes. Nature 2017, 542, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hille, F.; Charpentier, E. CRISPR-cas: Biology, mechanisms and relevance. Philos. Trans. R. Soc. B 2016, 371, 20150496. [Google Scholar] [CrossRef]
- Shiriaeva, A.; Fedorov, I.; Vyhovskyi, D.; Severinov, K. Detection of CRISPR adaptation. Biochem. Soc. Trans. 2020, 48, 257–269. [Google Scholar] [CrossRef] [Green Version]
- McGinn, J.; Marraffini, L.A. Molecular mechanisms of CRISPR–Cas spacer acquisition. Nat. Rev. Microbiol. 2019, 17, 7–12. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Didovik, A.; Bartłomiej, B.; Lev, T.; Hasty, J. Transcriptional Regulation with CRISPR-Cas9: Principles, Advances, and Applications. Curr. Opin. Biotechnol. 2016, 40, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Abid, H.Z.; Young, E.; McCaffrey, J.; Raseley, K.; Varapula, D.; Wang, H.Y.; Piazza, D.; Mell, J.; Xiao, M. Customized optical mapping by CRISPR-Cas9 mediated DNA labeling with multiple sgRNAs. Nucleic Acids Res. 2021, 49, e8. [Google Scholar] [CrossRef]
- Wang, X.; Shang, X.; Huang, X. Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerg. Microbes Infect. 2020, 9, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, S.; Lutz, C.; Henneman, L.; Bhin, J.; Wong, K.; Siteur, B.; van Gerwen, B.; de Korte-Grimmerink, R.; Zafra, M.P.; Schatoff, E.M.; et al. In situ CRISPR-Cas9 base editing for the development of genetically engi-neered mouse models of breast cancer. EMBO J. 2020, 39, e102169. [Google Scholar] [CrossRef]
- Liu, Z.; Liao, Z.; Chen, Y.; Zhou, L.; Huangting, W.; Xiao, H. Research on CRISPR/system in major cancers and its potential in cancer treatments. Clin. Transl. Oncol. 2021, 23, 425–433. [Google Scholar] [CrossRef]
- Wang, W.; Song, F.; Feng, X.; Chu, X.; Dai, H.; Tian, J.; Fang, X.; Song, F.; Liu, B.; Li, L.; et al. Functional Interrogation of Enhancer Connectome Prioritizes Candidate Target Genes at Ovarian Cancer Susceptibility Loci. Front. Genet. 2021, 12, 261. [Google Scholar]
- Li, J.; Yuan, S.; Norgard, R.J.; Yan, F.; Sun, Y.H.; Kim, I.K.; Merrell, A.J.; Sela, Y.; Jiang, Y.; Bhanu, N.V.; et al. Epigenetic and transcriptional control of the epidermal growth factor receptor regulates the tumor immune microenvironment in pancreatic cancer. Cancer Discov. 2021, 11, 736–753. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Ouyang, W.; Kang, B.; Han, X.; Xiong, Y.; Ding, R.; Li, Y.; Wang, F.; Huang, L.; Chen, L.; et al. Selective targeting of the oncogenic KRAS G12S mutant allele by CRISPR/Cas9 induces efficient tumor regression. Theranostics 2020, 10, 5137–5153. [Google Scholar] [CrossRef]
- Qiu, X.Y.; Zhu, L.Y.; Zhu, C.S.; Ma, J.X.; Hou, T.; Wu, X.M.; Xie, S.S.; Min, L.; Tan, D.A.; Zhang, D.Y.; et al. Highly Effective and Low-Cost MicroRNA Detection with CRISPR-Cas9. ACS Synth. Biol. 2018, 7, 807–813. [Google Scholar] [CrossRef]
- Ledford, H. Landmark CRISPR trial shows promise against deadly disease. Nature 2021. [Google Scholar] [CrossRef]
- Lim, G.B. Gene editing in patients with amyloidosis. Nat. Rev. Cardiol. 2021, 18, 611. [Google Scholar] [PubMed]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’Connell, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef]
- Shan, Y.; Zhou, X.; Huang, R.; Xing, D. High-Fidelity and Rapid Quantification of miRNA Combining crRNA Programmability and CRISPR/Cas13a trans-Cleavage Activity. Anal. Chem. 2019, 91, 5278–5285. [Google Scholar] [CrossRef]
- Osborn, M.J.; Bhardwaj, A.; Bingea, S.P.; Knipping, F.; Feser, C.J.; Lees, C.J.; Collins, D.P.; Steer, C.J.; Blazar, B.R.; Tolar, J. CRISPR/Cas9-Based Lateral Flow and Fluorescence Diagnostics. Bioengineering 2021, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.F. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018, 9, 5012. [Google Scholar] [CrossRef] [Green Version]
- Bandaru, S.; Tsuji, M.H.; Shimizu, Y.; Usami, K.; Lee, S.; Takei, N.K.; Yoshitome, K.; Nishimura, Y.; Otsuki, T.; Ito, T. Structure-based design of gRNA for Cas13. Sci. Rep. 2020, 10, 11610. [Google Scholar] [CrossRef]
- Wessels, H.H.; Méndez-Mancilla, A.; Guo, X.; Legut, M.; Daniloski, Z.; Sanjana, N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020, 38, 722–727. [Google Scholar] [CrossRef]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339. [Google Scholar] [CrossRef] [Green Version]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yin, L.; Dong, Y.; Peng, L.; Liu, G.; Man, S.; Ma, L. CRISPR-Cas13a based bacterial detection platform: Sensing pathogen Staphylococcus aureus in food samples. Anal. Chim. Acta 2020, 1127, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Baerwald, M.R.; Goodbla, A.M.; Nagarajan, R.P.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F.; Schreier, A.D. Rapid and accurate species identification for ecological studies and monitoring using CRISPR-based SHERLOCK. Mol. Ecol. Resour. 2020, 20, 961–970. [Google Scholar] [CrossRef]
- Durán-Vinet, B.; Araya-Castro, K.; Chao, T.C.; Wood, S.A.; Gallardo, V.; Godoy, K.; Abanto, M. Potential applications of CRISPR/Cas for next-generation biomonitoring of harmful algae blooms: A review. Harmful Algae 2021, 103, 102027. [Google Scholar] [CrossRef]
- Granados-Riveron, J.T.; Aquino-Jarquin, G. CRISPR/Cas13-Based Approaches for Ultrasensitive and Specific Detection of microRNAs. Cells 2021, 10, 1655. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Vinchhi, P.; Patel, M.M. Triumph against cancer: Invading colorectal cancer with nanotechnology. Expert Opin. Drug Deliv. 2021, 18, 1169–1192. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection using Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, H.; Taguchi, Y.; Nakagawa, R.; Makino, A.; Okazaki, S.; Nakano, M.; Muramoto, Y.; Takahashi, C.; Takahashi, I.; Ando, J.; et al. Amplification-free RNA detection with CRISPR–Cas13. Commun. Biol. 2021, 4, 476. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Huang, R.; Huang, M.; Yue, H.; Shan, Y.; Hu, J.; Xing, D. Cascade CRISPR/cas enables amplification-free microRNA sensing with fM-sensitivity and single-base-specificity. Chem. Commun. 2021, 57, 247–250. [Google Scholar] [CrossRef]
- Cui, Y.; Fan, S.; Yuan, Z.; Song, M.; Hu, J.; Qian, D.; Zhen, D.; Li, J.; Zhu, B. Ultrasensitive electrochemical assay for mi-croRNA-21 based on CRISPR/Cas13a-assisted catalytic hairpin assembly. Talanta 2021, 224, 121878. [Google Scholar] [CrossRef] [PubMed]
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, 1905311. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.; Huang, R.; Huang, M.; Shen, J.; Shan, Y.; Xing, D. CRISPR/Cas13a Powered Portable Electrochemiluminescence Chip for Ultrasensitive and Specific MiRNA Detection. Adv. Sci. 2020, 7, 1903661. [Google Scholar] [CrossRef] [PubMed]
- Wee, E.J.H.; Trau, M. Simple Isothermal Strategy for Multiplexed, Rapid, Sensitive, and Accurate miRNA Detection. ACS Sens. 2016, 1, 670–675. [Google Scholar] [CrossRef]
- Dunnett, H.; van der Meer, D.; Williams, G.A. Evaluation of stem-loop reverse transcription and poly-A tail extension in microRNA analysis of body fluids. Microrna 2014, 3, 150–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Kellner, M.J.; Zhang, F. Nucleic Acid Detection of Plant Genes Using CRISPR-Cas13. CRISPR J. 2019, 2, 165–171. [Google Scholar] [CrossRef] [Green Version]
- Addgene pC013-Twinstrep-SUMO-huLwCas13a (Plasmid #90097). Available online: https://www.addgene.org/90097/ (accessed on 2 June 2021).
- Ding, M.; Wang, C.; Lu, X.; Zhang, C.; Zhou, Z.; Chen, X.; Zhang, C.Y.; Zen, K.; Zhang, C. Comparison of commercial exosome isolation kits for circulating exosomal microRNA profiling. Anal. Bioanal. Chem. 2018, 410, 3805–3814. [Google Scholar] [CrossRef]




| Methods | Cost | Time * | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Traditional methods | |||||
| Guaiac-based fecal occult blood test (gFOBT) | Low | Weeks | Biennial gFOBT screening provides sustained protection against long-term CRC mortality | Unspecific, limited sensitivity for CRC detection, requires patient dietary modification and three consecutive samples are needed. | [73,74,75,76] |
| Fecal immunochemical test (FIT) | Low | Weeks | Quantitative and qualitative results, user-friendly application, higher overall adherence and easier follow-up. | Test reliability decreases considerably with longer times before analysis. | [74,75,76] |
| Multi-target stool DNA test | High | Weeks | High sensitivity, non-invasive approach, and good benefit-risk ratios | Sensitivity is partially linked to hemoglobin thresholds and showcases a lower rate of cancer prevention | [75,77] |
| Colonoscopy | High | Hours | High efficacy and sensitivity on preventing CRC due to detecting and removing both advanced and non-advanced adenomas. | Invasive, need sedation and bowel cleansing. High risks linked to human manipulation errors including perforation, bleeding, and death. | [75,77] |
| CT colonography (virtual colonoscopy) | High | Hours | Non-invasive and effective screening test with low risk of perforation | Bowel preparation and lower sensitivity in comparison with colonoscopy. | [75,77] |
| Molecular methods | |||||
| qPCR | Low | Days | Minimally invasive, fast, and accurate detection. The process has been automated. | Multi-target approaches and fluorescent reporters-related applicability is variable, affecting sensitivity and specificity. | [78,79] |
| RT-qPCR | Low | Days | Minimally invasive and accurate detection. Currently gold-standard method. | Error-prone and reliability directly linked to sample extraction quality from clinical samples. Labor-intensive. Low portability. | [78,80] |
| ddPCR | Low | Days | Minimally invasive with improved analytical sensitivity to mutations such as KRAS. Reduced variability. | Trained personnel, labor-intensive, and high rates of false positives. | [81,82] |
| Microarrays | Medium | Days-Weeks | Minimally invasive with high sensitivity to analyze multiple targets from one sample. | Time and laborious technical procedures, along with multiple runs needed to obtain final results. | [83] |
| Next-generation sequencing | High | Weeks | A broader assessment of the tumor molecular profile, including mutations and ITH dynamics. | Resource-consuming and efficacy may be affected by numerous factors | [84] |
| CRISPR/Cas platforms | Very low | Hours-Days | Minimally invasive detection with swift, cost-effective, ultrasensitive, and specific platforms. | Detailed sequence data needed, sensitive to unidentified mutations and RNA secondary structures. | [22,24,85] |
| Molecular Biomarkers | Sample Type a | Example Target | Overall Effectiveness (SE/SP) | References |
|---|---|---|---|---|
| Adenomatous polyposis coli (APC) | Blood (DNA) | D18122V, E1317Q, and I1307K (APC polymorphisms) | NR * | [89] |
| Microsatellite instability (MSI) | Blood (DNA) | Bat-25, NR-21 | 99% (98.7/100) | [90] |
| Methylation (MTL) | Blood/Stool (DNA) | SEPT9 | 89% (90/88) | [91] |
| Kirsten rat sarcoma viral oncogene homolog (KRAS) | Blood (DNA) | p-21Ras mutations | 60% (67/53.95) | [92] |
| V-raf murine sarcoma vViral oncogene homolog B1 (BRAF) | Blood (DNA) | BRAF V600 E mutation | 77% (81.2/72.1) | [93] |
| miRNA (By Stages) a | Qualitative Regulation b | AUC | Sequence (3p/5p-length-bp) c | Accession Number | Qualitative Prognosis | Source | Reference |
|---|---|---|---|---|---|---|---|
| T (I & II) | |||||||
| miR-126 | ↑ | 0.96 | UCG UAC CGU GAG UAA UAA UGC G (3p-22) | MI0000471 | Early CRC stage | CEx | [136] |
| miR-1290 | ↑ | 0.91 | UGG AUU UUU GGA UCA GGG A (19) | MI0006352 | Early CRC stage | CEx | [136] |
| miR-186-5p | ↑ | 0.72 | CAAA GAA UUC UCC UUU UGG GCU (21) | MI0000483 | CRC early lesions | cf-miRNAs | [139] |
| miR-23a | ↑ | 0.92 | AUC ACA UUG CCA GGG AUU UCC (3p-21) | MI0000079 | Early CRC stage | CEx | [136] |
| miR-423-5p | ↓ | 0.72 | UGA GGG GCA GAG AGC GAG ACU UU (23) | MI0001445 | CRC early lesions | cf-miRNAs | [139] |
| miR-449a | ↓ | 0.76 | UGG CAG UGU AUU GUU AGC UGG U (22) | MI0001648 | Poor prognosis, lower overall survival | cf-miRNAs | [140] |
| miR-592 | ↑ | 0.80 | UUG UGU CAA UAU GCG AUG AUG U (22) | MI0003604 | Early CRC stage | cf-miRNAs | [141] |
| miR-940 | ↑ | 0.90 | AAG GCA GGG CCC CCG CUC CCC (21) | MI0005762 | Early CRC stage | CEx | [136] |
| N (III) | |||||||
| miR-1539 | ↑ | 0.67 | UCC UGC GCG UCC CAG AUG CCC (21) | MI0007260 | CRC lymph node invasion and poor clinicopathological behavior | CEx | [142] |
| miR-19a | ↑ | 0.87 | UGU GCA AAU CUA UGC AAA ACU GA (3p-23) | MI0000073 | CRC invasion | cf-miRNAs | [143] |
| miR-20a | ↑ | 0.83 | UAA AGU GCU UAU AGU GCA GGU AG (5p-23) | MI0000076 | CRC increasing distant metastasis rates | cf-miRNAs | [143] |
| miR-150 | ↑ | 0.75 | UCU CCC AAC CCU UGU ACC AGU G (5p-22) | MI0000479 | CRC promoting epithelial to mesenchymal transition | cf-miRNAs | [143] |
| miR-552 | ↑ | NR | AAC AGG UGA CUG GUU AGA CAA (3p-21) | MI0003557 | CRC poor prognosis, worse 5-year overall survival | cf-miRNAs | [144] |
| M (IV) | |||||||
| miR-126-3p | ↑ | NR | UCG UAC CGU GAG UAA UAA UGC G (22) | MI0000471 | *Progression-free survival | cf-miRNAs | [145] |
| miR-155-5p | ↑ | NR | UUA AUG CUA AUC GUG AUA GGG GUU (24) | MI0000681 | *Short progression-free survival | cf-miRNAs | [146] |
| miR-17-5p | ↑ | 0.90 | CAA AGU GCU UAC AGU GCA GGU AG (23) | MI0000071 | CRC increased invasive ability and metastasis potential | CEx | [147] |
| miR-19b | ↑ | 0.89 | UGU GCA AAU CCA UGC AAA ACU GA (3p 23) | MI0000074 | High amounts indicate metastatic CRC | CEx | [132] |
| miR-20b-5p | ↑ | NR | CAA AGU GCU CAU AGU GCA GGU AG (23) | MI0001519 | *Progression-free survival | cf-miRNAs | [146] |
| miR-21 | ↑ | 0.98 | UAG CUU AUC AGA CUG AUG UUG A (5p-22) | MI0000077 | High amounts indicate metastatic CRC | CEx | [132] |
| miR-222 | ↑ | 0.90 | AGC UAC AUC UGG CUA CUG GGU (3p-22) | MI0000299 | Higher amounts indicate a lower overall survival rate | CEx | [132] |
| miR-29b-3p | ↑ | NR | UAG CAC CAU UUG AAA UCA GUG UU (23) | MI0000105 | *Progression-free survival | cf-miRNAs | [146] |
| miR-320d | ↑ | 0.63 | AAA AGC UGG GUU GAG AGG A (19) | MI0008190 | Distinguish metastatic from non-metastatic CRC. | CEx | [148] |
| miR-92a | ↑ | 0.95 | UAU UGC ACU UGU CCC GGC CUG U (3p-22) | MI0000093 | Higher amounts indicate a higher risk of tumor progression | CEx | [132] |
| miR-92a-3p | ↑ | 0.85 | UAU UGC ACU UGU CCC GGC CUG U (22) | MI0000093 | CRC increased invasive ability and metastasis potential | CEx | [147] |
| Methods a | Target | Pre-Amplification (Method) | Sensitivity b | Runtime (min) | Multiplexation | Readout | Reference |
|---|---|---|---|---|---|---|---|
| miRNA targets approach | |||||||
| CRISPR/LbuCas13a | miRNAs | N | 5 pM | 30 | N | F | [179] |
| non-miRNA targets approach | |||||||
| SHERLOCK | ST | Y (RPA) | 2 aM | 120 | N | F | [21] |
| SHERLOCKv2 | ST | Y (RPA) | 8 zM | 30 | Y (4) | F/S | [22] |
| HUDSON | ST | Y (RPA) | 0.9 aM | 120 | N | F/S | [26] |
| CARMEN-Cas13 | Viral particles | Y (PCR) | 2 aM | 30–180 | Y (169) | F | [192] |
| SATORI | SARS-CoV-2 | N | 5 fM | 5–10 | N | F | [193] |
| miRNA | crRNA Sequence a | Cas13 Ortholog (Spacer Length) | Associated FQR | Reference |
|---|---|---|---|---|
| Multiplex approach b | ||||
| miR-126-3p | {GAU UUA GAC UAC CCC AAA AAC GAA GGG GAC UAA AAC}–[AGC AUG GCA CUC AUU AUU ACG C (uuu uuu)] | LwaCas13a (28 nt) | F//T*A*rArUG*C//Q | [24,136,197] |
| miR-1290 | {GUU GAU GAG AAG AGC CCA AGA UAG AGG GCA AUA AC}–[ACC UAA AAA CCU AGU CCC U (uuu uuu uuu)] | LbaCas13a (28 nt) | F//T*A*rUrAC*C*//Q | [24,136,197] |
| miR-23a-3p | [UAG UGU AAC GGU CCC UAA AGG (uuu uuu uuu)]–{GUU GUA GAA GCU UAU CGU UUG GAU AGG UAU GAC AAC} | CcaCas13b (30 nt) | F//T*A*rUrAG*C*//Q | [24,136,197] |
| miR-940 | [UUC CGU CCC GGG GGC GAG GGG (uuu uuu uuu)]–{GUU GUA GAA GCU UAU CGU UUG GAU AGG UAU GAC AAC} | PsmCas13b (30 nt) | F//rArArArArA//Q | [24,136,197] |
| Singleplex approach c | ||||
| miR-126-3p | {GAU UUA GAC UAC CCC AAA AAC GAA GGG GAC UAA AAC}–[AGC AUG GCA CUC AUU AUU ACG C (uuu uuu)] | LwaCas13a (28 nt) | F//T*A*rArUG*C//Q | [24,136,197] |
| miR-1290 | {GAU UUA GAC UAC CCC AAA AAC GAA GGG GAC UAA AAC}–[ACC UAA AAA CCU AGU CCC U (uuu uuu uuu)] | LwaCas13a (28 nt) | F//T*A*rArUG*C//Q | [24,136,197] |
| miR-23a-3p | {GAU UUA GAC UAC CCC AAA AAC GAA GGG GAC UAA AAC}–[UAG UGU AAC GGU CCC UAA AGG (uuu uuu u)] | LwaCas13a (28 nt) | F//T*A*rArUG*C//Q | [24,136,197] |
| miR-940 | {GAU UUA GAC UAC CCC AAA AAC GAA GGG GAC UAA AAC}–[UUC CGU CCC GGG GGC GAG GGG (uuu uuu u)] | LwaCas13a (28 nt) | F//T*A*rArUG*C//Q | [24,136,197] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durán-Vinet, B.; Araya-Castro, K.; Calderón, J.; Vergara, L.; Weber, H.; Retamales, J.; Araya-Castro, P.; Leal-Rojas, P. CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review. Cancers 2021, 13, 4640. https://doi.org/10.3390/cancers13184640
Durán-Vinet B, Araya-Castro K, Calderón J, Vergara L, Weber H, Retamales J, Araya-Castro P, Leal-Rojas P. CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review. Cancers. 2021; 13(18):4640. https://doi.org/10.3390/cancers13184640
Chicago/Turabian StyleDurán-Vinet, Benjamín, Karla Araya-Castro, Juan Calderón, Luis Vergara, Helga Weber, Javier Retamales, Paulina Araya-Castro, and Pamela Leal-Rojas. 2021. "CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review" Cancers 13, no. 18: 4640. https://doi.org/10.3390/cancers13184640
APA StyleDurán-Vinet, B., Araya-Castro, K., Calderón, J., Vergara, L., Weber, H., Retamales, J., Araya-Castro, P., & Leal-Rojas, P. (2021). CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review. Cancers, 13(18), 4640. https://doi.org/10.3390/cancers13184640