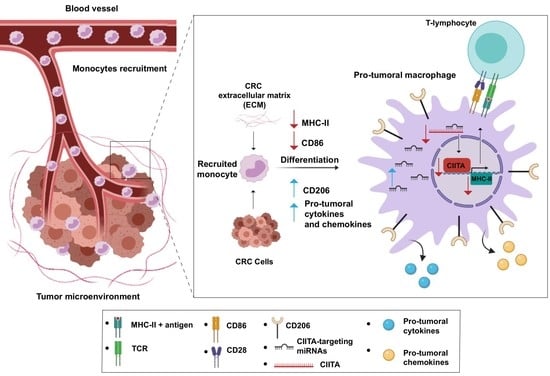
Tumor Cells and the Extracellular Matrix Dictate the Pro-Tumoral Profile of Macrophages in CRC
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Patients
2.3. Preparation of Decellularized Matrices
2.4. Cell Cultures
2.4.1. Tumor and Normal Intestinal Cells
2.4.2. Conditioned Media Preparation
2.4.3. Monocyte Isolation
2.4.4. Co-Culture of Monocytes and Intestinal Cells
2.4.5. Co-Culture of Monocytes and Decellularized Matrices
2.5. Immunohistochemistry
2.6. Flow Cytometry
2.7. RNA Extraction
2.8. qRT-PCR
2.8.1. mRNAs
2.8.2. miRNAs
2.9. Quantification of Cytokines, Chemokines, and Hyaluronic Acid (HA) in Culture Supernatants
2.9.1. ELISA
2.9.2. Multiplex
2.9.3. HA Quantification
2.10. T Cell Proliferation Assay
2.11. Statistics
3. Results
3.1. Macrophages Expressing Low MHC-II and High CD206 Infiltrate CRC Mucosa
3.2. Tumor Cells Educate Monocytes to Acquire a Pro-Tumoral Macrophage-like Profile
3.3. Tumor Cells Induce the Production of Anti-Inflammatory and Pro-Tumoral Cytokines and Chemokines in Monocyte-Differentiated Macrophages
3.4. The Extracellular Matrix Educates Monocytes to Acquire a Pro-Tumoral Macrophage-like Profile
3.5. The Down-Modulation of MHC-II Involves the Targeting of CIITA by miR146b and let-7i
3.6. Decreased MHC-II Expression Affects the Activation of a Specific T Cell Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Patterns and Trends in Colorectal Cancer Incidence and Mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Mantovani, A.; Ponzetta, A.; Inforzato, A.; Jaillon, S. Innate Immunity, Inflammation and Tumour Progression: Double-Edged Swords. J. Intern. Med. 2019, 285, 524–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, B.-Z.; Pollard, J.W. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Sica, A.; Locati, M. Macrophage Polarization Comes of Age. Immunity 2005, 23, 344–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sica, A.; Erreni, M.; Allavena, P.; Porta, C. Macrophage Polarization in Pathology. Cell. Mol. Life Sci. 2015, 72, 4111–4126. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.K.; Chittezhath, M.; Shalova, I.N.; Lim, J.-Y. Macrophage Polarization and Plasticity in Health and Disease. Immunol. Res. 2012, 53, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Solinas, G.; Germano, G.; Mantovani, A.; Allavena, P. Tumor-Associated Macrophages (TAM) as Major Players of the Cancer-Related Inflammation. J. Leukoc. Biol. 2009, 86, 1065–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaput, N.; Svrcek, M.; Aupérin, A.; Locher, C.; Drusch, F.; Malka, D.; Taïeb, J.; Goéré, D.; Ducreux, M.; Boige, V. Tumour-Infiltrating CD68+ and CD57+ Cells Predict Patient Outcome in Stage II–III Colorectal Cancer. Br. J. Cancer 2013, 109, 1013–1022. [Google Scholar] [CrossRef] [Green Version]
- Koelzer, V.H.; Canonica, K.; Dawson, H.; Sokol, L.; Karamitopoulou-Diamantis, E.; Lugli, A.; Zlobec, I. Phenotyping of Tumor-Associated Macrophages in Colorectal Cancer: Impact on Single Cell Invasion (Tumor Budding) and Clinicopathological Outcome. OncoImmunology 2016, 5, e1106677. [Google Scholar] [CrossRef] [Green Version]
- Ålgars, A.; Irjala, H.; Vaittinen, S.; Huhtinen, H.; Sundström, J.; Salmi, M.; Ristamäki, R.; Jalkanen, S. Type and Location of Tumor-Infiltrating Macrophages and Lymphatic Vessels Predict Survival of Colorectal Cancer Patients. Int. J. Cancer 2012, 131, 864–873. [Google Scholar] [CrossRef]
- Gulubova, M.; Ananiev, J.; Yovchev, Y.; Julianov, A.; Karashmalakov, A.; Vlaykova, T. The Density of Macrophages in Colorectal Cancer Is Inversely Correlated to TGF-Β1 Expression and Patients’ Survival. J. Mol. Hist. 2013, 44, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Peng, R.-Q.; Wu, X.-J.; Xia, Q.; Hou, J.-H.; Ding, Y.; Zhou, Q.-M.; Zhang, X.; Pang, Z.-Z.; Wan, D.-S.; et al. The Density of Macrophages in the Invasive Front Is Inversely Correlated to Liver Metastasis in Colon Cancer. J. Transl. Med. 2010, 8, 13. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.-L.; Li, H.-K.; Zhou, H.-Y.; Zhang, T.; Li, Q. Correlations of Tumor-Associated Macrophage Subtypes with Liver Metastases of Colorectal Cancer. Asian Pac. J. Cancer Prev. APJCP 2013, 14, 1003–1007. [Google Scholar] [CrossRef] [Green Version]
- Hamm, A.; Prenen, H.; Van Delm, W.; Di Matteo, M.; Wenes, M.; Delamarre, E.; Schmidt, T.; Weitz, J.; Sarmiento, R.; Dezi, A.; et al. Tumour-Educated Circulating Monocytes Are Powerful Candidate Biomarkers for Diagnosis and Disease Follow-up of Colorectal Cancer. Gut 2016, 65, 990–1000. [Google Scholar] [CrossRef]
- Franklin, R.A.; Liao, W.; Sarkar, A.; Kim, M.V.; Bivona, M.R.; Liu, K.; Pamer, E.G.; Li, M.O. The Cellular and Molecular Origin of Tumor-Associated Macrophages. Science 2014, 344, 921–925. [Google Scholar] [CrossRef] [Green Version]
- Väyrynen, J.P.; Haruki, K.; Lau, M.C.; Väyrynen, S.A.; Zhong, R.; Dias Costa, A.; Borowsky, J.; Zhao, M.; Fujiyoshi, K.; Arima, K.; et al. The Prognostic Role of Macrophage Polarization in the Colorectal Cancer Microenvironment. Cancer Immunol. Res. 2021, 9, 8–19. [Google Scholar] [CrossRef]
- Pinto, M.L.; Rios, E.; Durães, C.; Ribeiro, R.; Machado, J.C.; Mantovani, A.; Barbosa, M.A.; Carneiro, F.; Oliveira, M.J. The Two Faces of Tumor-Associated Macrophages and Their Clinical Significance in Colorectal Cancer. Front. Immunol. 2019, 10, 1875. [Google Scholar] [CrossRef] [Green Version]
- Accolla, R.S.; Tosi, G. Optimal MHC-II-Restricted Tumor Antigen Presentation to CD4+ T Helper Cells: The Key Issue for Development of Anti-Tumor Vaccines. J. Transl. Med. 2012, 10, 154. [Google Scholar] [CrossRef] [Green Version]
- Reeves, E.; James, E. Antigen Processing and Immune Regulation in the Response to Tumours. Immunology 2017, 150, 16–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandola-Simon, J.; Roche, P.A. Dysfunction of Antigen Processing and Presentation by Dendritic Cells in Cancer. Mol. Immunol. 2019, 113, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Warabi, M.; Kitagawa, M.; Hirokawa, K. Loss of MHC Class II Expression Is Associated with a Decrease of Tumor-Infiltrating T Cells and an Increase of Metastatic Potential of Colorectal Cancer: Immunohistological and Histopathological Analyses as Compared with Normal Colonic Mucosa and Adenomas. Pathol. Res. Pract. 2000, 196, 807–815. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.-S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International Validation of the Consensus Immunoscore for the Classification of Colon Cancer: A Prognostic and Accuracy Study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Matsushita, K.; Takenouchi, T.; Shimada, H.; Tomonaga, T.; Hayashi, H.; Shioya, A.; Komatsu, A.; Matsubara, H.; Ochiai, T. Strong HLA-DR Antigen Expression on Cancer Cells Relates to Better Prognosis of Colorectal Cancer Patients: Possible Involvement of c-Myc Suppression by Interferon-Gammain Situ. Cancer Sci. 2006, 97, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.; Rognum, T.; Lund, E.; Meling, G.; Hauge, S. Strong HLA-DR Expression in Large Bowel Carcinomas Is Associated with Good Prognosis. Br. J. Cancer 1993, 68, 80–85. [Google Scholar] [CrossRef]
- D’Angelo, E.; Natarajan, D.; Sensi, F.; Ajayi, O.; Fassan, M.; Mammano, E.; Pilati, P.; Pavan, P.; Bresolin, S.; Preziosi, M.; et al. Patient-Derived Scaffolds of Colorectal Cancer Metastases as an Organotypic 3D Model of the Liver Metastatic Microenvironment. Cancers 2020, 12, 364. [Google Scholar] [CrossRef] [Green Version]
- Kuang, D.-M.; Wu, Y.; Chen, N.; Cheng, J.; Zhuang, S.-M.; Zheng, L. Tumor-Derived Hyaluronan Induces Formation of Immunosuppressive Macrophages through Transient Early Activation of Monocytes. Blood 2007, 110, 587–595. [Google Scholar] [CrossRef]
- D’Elios, M.M.; Vallese, F.; Capitani, N.; Benagiano, M.; Bernardini, M.L.; Rossi, M.; Rossi, G.P.; Ferrari, M.; Baldari, C.T.; Zanotti, G.; et al. The Helicobacter Cinaedi Antigen CAIP Participates in Atherosclerotic Inflammation by Promoting the Differentiation of Macrophages in Foam Cells. Sci. Rep. 2017, 7, 40515. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/Macrosialin: Not Just a Histochemical Marker. Lab. Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef] [Green Version]
- Pagliari, M.; Munari, F.; Toffoletto, M.; Lonardi, S.; Chemello, F.; Codolo, G.; Millino, C.; DELLA Bella, C.; Pacchioni, B.; Vermi, W.; et al. Helicobacter pylori Affects the Antigen Presentation Activity of Macrophages Modulating the Expression of the Immune Receptor CD300E through miR-4270. Front. Immunol. 2017, 8, 1288. [Google Scholar] [CrossRef] [Green Version]
- D’Elios, M.M.; Josien, R.; Manghetti, M.; Amedei, A.; De Carli, M.; Cuturi, M.C.; Blancho, G.; Buzelin, F.; Del Prete, G.; Soulillou, J.P. Predominant Th1 Cell Infiltration in Acute Rejection Episodes of Human Kidney Grafts. Kidney Int. 1997, 51, 1876–1884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erreni, M.; Mantovani, A.; Allavena, P. Tumor-Associated Macrophages (TAM) and Inflammation in Colorectal Cancer. Cancer Microenviron. Off. J. Int. Cancer Microenviron. Soc. 2011, 4, 141–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Rajput, A.; Jin, N.; Wang, J. Mechanisms of Immunosuppression in Colorectal Cancer. Cancers 2020, 12, 3850. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Weaver, V.M.; Werb, Z. The Extracellular Matrix: A Dynamic Niche in Cancer Progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.; Mojares, E.; del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. IJMS 2018, 19, 3028. [Google Scholar] [CrossRef] [Green Version]
- Song, J.J.; Ott, H.C. Organ Engineering Based on Decellularized Matrix Scaffolds. Trends Mol. Med. 2011, 17, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Hoshiba, T.; Tanaka, M. Breast Cancer Cell Behaviors on Staged Tumorigenesis-Mimicking Matrices Derived from Tumor Cells at Various Malignant Stages. Biochem. Biophys. Res. Commun. 2013, 439, 291–296. [Google Scholar] [CrossRef]
- Piccoli, M.; D’Angelo, E.; Crotti, S.; Sensi, F.; Urbani, L.; Maghin, E.; Burns, A.; De Coppi, P.; Fassan, M.; Rugge, M.; et al. Decellularized Colorectal Cancer Matrix as Bioactive Microenvironment for in Vitro 3D Cancer Research. J. Cell. Physiol. 2018, 233, 5937–5948. [Google Scholar] [CrossRef]
- Sensi, F.; D’Angelo, E.; Piccoli, M.; Pavan, P.; Mastrotto, F.; Caliceti, P.; Biccari, A.; Corallo, D.; Urbani, L.; Fassan, M.; et al. Recellularized Colorectal Cancer Patient-Derived Scaffolds as In Vitro Pre-Clinical 3D Model for Drug Screening. Cancers 2020, 12, 681. [Google Scholar] [CrossRef] [Green Version]
- Pinto, M.L.; Rios, E.; Silva, A.C.; Neves, S.C.; Caires, H.R.; Pinto, A.T.; Durães, C.; Carvalho, F.A.; Cardoso, A.P.; Santos, N.C.; et al. Decellularized Human Colorectal Cancer Matrices Polarize Macrophages towards an Anti-Inflammatory Phenotype Promoting Cancer Cell Invasion via CCL18. Biomaterials 2017, 124, 211–224. [Google Scholar] [CrossRef]
- Boss, J.M. Regulation of Transcription of MHC Class II Genes. Curr. Opin. Immunol. 1997, 9, 107–113. [Google Scholar] [CrossRef]
- Reith, W.; LeibundGut-Landmann, S.; Waldburger, J.-M. Regulation of MHC Class II Gene Expression by the Class II Transactivator. Nat. Rev. Immunol. 2005, 5, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Holling, T.M.; Schooten, E.; Langerak, A.W.; van den Elsen, P.J. Regulation of MHC Class II Expression in Human T-Cell Malignancies. Blood 2004, 103, 1438–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, S.P.; Tomasi, T.B. Absence of MHC Class II Antigen Expression in Trophoblast Cells Results from a Lack of Class II Transactivator (CIITA) Gene Expression. Mol. Reprod. Dev. 1998, 51, 1–12. [Google Scholar] [CrossRef]
- Morimoto, Y.; Toyota, M.; Satoh, A.; Murai, M.; Mita, H.; Suzuki, H.; Takamura, Y.; Ikeda, H.; Ishida, T.; Sato, N.; et al. Inactivation of Class II Transactivator by DNA Methylation and Histone Deacetylation Associated with Absence of HLA-DR Induction by Interferon-γ in Haematopoietic Tumour Cells. Br. J. Cancer 2004, 90, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Croce, C.M.; Calin, G.A. MicroRNAs. Cancer J. 2008, 14, 1–6. [Google Scholar] [CrossRef]
- Codolo, G.; Toffoletto, M.; Chemello, F.; Coletta, S.; Soler Teixidor, G.; Battaggia, G.; Munari, G.; Fassan, M.; Cagnin, S.; de Bernard, M. Helicobacter Pylori Dampens HLA-II Expression on Macrophages via the Up-Regulation of MiRNAs Targeting CIITA. Front. Immunol. 2020, 10, 2923. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.O.; Kim, E.K.; Lee, M.; Jung, W.Y.; Lee, H.; Kang, Y.; Jang, Y.-J.; Hong, S.W.; Choi, S.H.; Yang, W.I. NOVA1 Inhibition by MiR-146b-5p in the Remnant Tissue Microenvironment Defines Occult Residual Disease after Gastric Cancer Removal. Oncotarget 2016, 7, 2475–2495. [Google Scholar] [CrossRef]
- Deng, X.; Wu, B.; Xiao, K.; Kang, J.; Xie, J.; Zhang, X.; Fan, Y. MiR-146b-5p Promotes Metastasis and Induces Epithelial-Mesenchymal Transition in Thyroid Cancer by Targeting ZNRF3. Cell Physiol. Biochem. 2015, 35, 71–82. [Google Scholar] [CrossRef]
- Zhu, Y.; Wu, G.; Yan, W.; Zhan, H.; Sun, P. MiR-146b-5p Regulates Cell Growth, Invasion, and Metabolism by Targeting PDHB in Colorectal Cancer. Am. J. Cancer Res. 2017, 7, 1136–1150. [Google Scholar]
- Zhang, P.; Ma, Y.; Wang, F.; Yang, J.; Liu, Z.; Peng, J.; Qin, H. Comprehensive gene and microRNA expression profiling reveals the crucial role of hsa-let-7i and its target genes in colorectal cancer metastasis. Mol. Biol. Rep. 2011, 39, 1471–1478. [Google Scholar] [CrossRef]
- Baer, C.; Squadrito, M.L.; Laoui, D.; Thompson, D.; Hansen, S.K.; Kiialainen, A.; Hoves, S.; Ries, C.H.; Ooi, C.-H.; De Palma, M. Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nature 2016, 18, 790–802. [Google Scholar] [CrossRef]
- Kryczek, I.; Zou, L.; Rodriguez, P.; Zhu, G.; Wei, S.; Mottram, P.; Brumlik, M.; Cheng, P.; Curiel, T.; Myers, L.; et al. B7-H4 Expression Identifies a Novel Suppressive Macrophage Population in Human Ovarian Carcinoma. J. Exp. Med. 2006, 203, 871–881. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Cunha, B.R.; Domingos, C.; Stefanini, A.C.B.; Henrique, T.; Polachini, G.M.; Castelo-Branco, P.; Tajara, E.H. Cellular Interactions in the Tumor Microenvironment: The Role of Secretome. J. Cancer 2019, 10, 4574–4587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardighieri, L.; Missale, F.; Bugatti, M.; Gatta, L.B.; Pezzali, I.; Monti, M.; Gottardi, S.; Zanotti, L.; Bignotti, E.; Ravaggi, A.; et al. Infiltration by CXCL10 Secreting Macrophages Is Associated With Antitumor Immunity and Response to Therapy in Ovarian Cancer Subtypes. Front. Immunol. 2021, 12, 690201. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Mantovani, A. Macrophage Plasticity and Interaction with Lymphocyte Subsets: Cancer as a Paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef]
- Knüpfer, H.; Preiss, R. Serum Interleukin-6 Levels in Colorectal Cancer Patients—a Summary of Published Results. Int. J. Colorectal. Dis. 2010, 25, 135–140. [Google Scholar] [CrossRef]
- Denning, T.L.; Wang, Y.; Patel, S.R.; Williams, I.R.; Pulendran, B. Lamina Propria Macrophages and Dendritic Cells Differentially Induce Regulatory and Interleukin 17–Producing T Cell Responses. Nat. Immunol. 2007, 8, 1086–1094. [Google Scholar] [CrossRef]
- Orsini, G.; Legitimo, A.; Failli, A.; Ferrari, P.; Nicolini, A.; Spisni, R.; Miccoli, P.; Consolini, R. Defective Generation and Maturation of Dendritic Cells from Monocytes in Colorectal Cancer Patients during the Course of Disease. IJMS 2013, 14, 22022–22041. [Google Scholar] [CrossRef] [Green Version]
- García-Lora, A.; Algarra, I.; Collado, A.; Garrido, F. Tumour Immunology, Vaccination and Escape Strategies: HLA and the Tumour Immune Escape. Eur. J. Immunogenet. 2003, 30, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-D.; Chu, C.-H.; Tsou, A.-P.; Chen, S.-J.; Chen, H.-C.; Hsu, P.W.-C.; Wong, Y.-H.; Chen, Y.-H.; Chen, G.-H.; Huang, H.-D. MiRNAMap 2.0: Genomic Maps of MicroRNAs in Metazoan Genomes. Nucleic Acids Res. 2007, 36, D165–D169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monslow, J.; Govindaraju, P.; Puré, E. Hyaluronan—A Functional and Structural Sweet Spot in the Tissue Microenvironment. Front. Immunol. 2015, 6, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwertfeger, K.L.; Cowman, M.K.; Telmer, P.G.; Turley, E.A.; McCarthy, J.B. Hyaluronan, Inflammation, and Breast Cancer Progression. Front. Immunol. 2015, 6, 236. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Cha, J.; Jang, M.; Kim, P. Hyaluronic Acid-Based Extracellular Matrix Triggers Spontaneous M2-like Polarity of Monocyte/Macrophage. Biomater. Sci. 2019, 7, 2264–2271. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Itano, N. Hyaluronan: A Modulator of the Tumor Microenvironment. Cancer Lett. 2016, 375, 20–30. [Google Scholar] [CrossRef]
- Sato, N.; Kohi, S.; Hirata, K.; Goggins, M. Role of Hyaluronan in Pancreatic Cancer Biology and Therapy: Once Again in the Spotlight. Cancer Sci. 2016, 107, 569–575. [Google Scholar] [CrossRef]
- Ropponen, K.; Tammi, M.; Parkkinen, J.; Eskelinen, M.; Tammi, R.; Lipponen, P.; Agren, U.; Alhava, E.; Kosma, V.M. Tumor Cell-Associated Hyaluronan as an Unfavorable Prognostic Factor in Colorectal Cancer. Cancer Res. 1998, 58, 342–347. [Google Scholar] [PubMed]
- Zhang, G.; Guo, L.; Yang, C.; Liu, Y.; He, Y.; Du, Y.; Wang, W.; Gao, F. A Novel Role of Breast Cancer-Derived Hyaluronan on Inducement of M2-like Tumor-Associated Macrophages Formation. OncoImmunology 2016, 5, e1172154. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| CRC Patients (n = 8) | ||
|---|---|---|
| Age | Median (range), yrs | 66 (58–88) |
| Sex | Male | 6 (75%) |
| Female | 2 (25%) | |
| TNM | I | 0 (0%) |
| II | 5 (62.5) | |
| III | 3 (37.5) | |
| IV | 0 (0%) | |
| Microsatellite status | Stable | 6 (75%) |
| Unstable | 1 (12.5%) | |
| Not available | 1 (12.5%) | |
| Grade | 1 | 0 (0%) |
| 2 | 6 (75%) | |
| 3 | 2 (25%) | |
| 4 | 0 (0%) | |
| Tumor histotype | Adenocarcinoma | 6 (75%) |
| Mucinous | 2 (25%) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coletta, S.; Lonardi, S.; Sensi, F.; D’Angelo, E.; Fassan, M.; Pucciarelli, S.; Valzelli, A.; Biccari, A.; Vermi, W.; Della Bella, C.; et al. Tumor Cells and the Extracellular Matrix Dictate the Pro-Tumoral Profile of Macrophages in CRC. Cancers 2021, 13, 5199. https://doi.org/10.3390/cancers13205199
Coletta S, Lonardi S, Sensi F, D’Angelo E, Fassan M, Pucciarelli S, Valzelli A, Biccari A, Vermi W, Della Bella C, et al. Tumor Cells and the Extracellular Matrix Dictate the Pro-Tumoral Profile of Macrophages in CRC. Cancers. 2021; 13(20):5199. https://doi.org/10.3390/cancers13205199
Chicago/Turabian StyleColetta, Sara, Silvia Lonardi, Francesca Sensi, Edoardo D’Angelo, Matteo Fassan, Salvatore Pucciarelli, Arianna Valzelli, Andrea Biccari, William Vermi, Chiara Della Bella, and et al. 2021. "Tumor Cells and the Extracellular Matrix Dictate the Pro-Tumoral Profile of Macrophages in CRC" Cancers 13, no. 20: 5199. https://doi.org/10.3390/cancers13205199
APA StyleColetta, S., Lonardi, S., Sensi, F., D’Angelo, E., Fassan, M., Pucciarelli, S., Valzelli, A., Biccari, A., Vermi, W., Della Bella, C., Barizza, A., D’Elios, M. M., de Bernard, M., Agostini, M., & Codolo, G. (2021). Tumor Cells and the Extracellular Matrix Dictate the Pro-Tumoral Profile of Macrophages in CRC. Cancers, 13(20), 5199. https://doi.org/10.3390/cancers13205199