Application of High-Z Gold Nanoparticles in Targeted Cancer Radiotherapy—Pharmacokinetic Modeling, Monte Carlo Simulation and Radiobiological Effect Modeling
Abstract
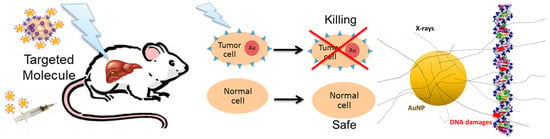
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Biological Datasets
2.1.1. Conjugation of AuNPs with cmHsp70.1 mAb
2.1.2. Analysis of AuNP Uptake
2.2. Pharmacokinetic Models
2.2.1. Model for SPIONs
2.2.2. Proposed Compartmental Model for Biodistribution of Targeted AuNPs in Cells
2.3. Gold Nanoparticle Based Spectral Computed Tomographic Imaging
2.4. Monte Carlo Simulation of Enhanced Radiosensitization Effect
2.4.1. Spectra of X-rays Used in Simulations
2.4.2. Dose Enhancement by a Single AuNP
2.4.3. Method for Dose and DNA Strand Breaks for Multiple AuNPs in Cells
2.4.4. Multi-Scale Method for Dose, Chemical Radical Generation and DNA Strand Breaks for Multiple AuNPs in a Xenograft Tumor
2.5. Mathematical Modeling of Enhanced Radiobiological Effect
3. Results
3.1. Visualization of AuNP Conjugates
3.1.1. Bright-Field and TEM Imaging
3.1.2. Spectral CT Imaging
3.2. Pharmacokinetic Model Developed for SPIONs
3.3. Simulated Dose and Biological Enhancement
3.3.1. Dose Enhancement Ratio for a Single AuNP
3.3.2. Enhancement Effect of Multiple AuNPs in Cellular Models
3.3.3. Enhancement Effect of Multiple AuNPs in Xenograft Tumor Model
3.4. Modeled Enhanced Radiobiological Effect—Cell Survival Fraction
4. Discussion
- Dose enhancement of a single AuNP in liquid water irradiated by a broad beam. These values might be used for the dose enhancement of multiple AuNPs in liquid water.
- Evaluation of the radiation-induced chemical radicals contributing to the enhancement effects of multiple AuNPs to the biological effects with advanced chemical module-implemented MC simulations.
- The cellular or molecular compartmental model for the heterogeneous uptake and distribution of AuNP-conjugates on the cell membrane and in the cytoplasm.
- Radiobiological experimental setup and defined endpoint results with injections of a controlled amounts of AuNPs with known shape composition and surface modifications, in vivo.
- Further advanced optical imaging technique for initial dynamic process of AuNPs in cells prior to and after irradiation.
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halperin, E.C.; Wazer, D.E.; Perez, C.A.; Brady, L.W. Perez and Brady’s Principles and Practice of Radiation Oncology, 6th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Iwamoto, K.S.; Cochran, S.T.; Winter, J.; Holburt, E.; Higashida, R.T.; Norman, A. Radiation dose enhancement therapy with iodine in rabbit VX-2 brain tumors. Radiother. Oncol. 1987, 8, 161–170. [Google Scholar] [CrossRef]
- Mesa, A.V.; Norman, A.; Solberg, T.D.; Demarco, J.J.; Smathers, J.B. Dose distributions using kilovoltage x-rays and dose enhancement from iodine contrast agents. Phys. Med. Biol. 1999, 44, 1955–1968. [Google Scholar] [CrossRef] [PubMed]
- Robar, J.L.; Riccio, S.A.; Martin, M.A. Tumour dose enhancement using modified megavoltage photon beams and contrast media. Phys. Med. Biol. 2002, 47, 2433–2449. [Google Scholar] [CrossRef]
- Solberg, T.D.; Iwamoto, K.S.; Norman, A. Calculation of radiation dose enhancement factors for dose enhancement therapy of brain tumours. Phys. Med. Biol. 1992, 37, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Cole, L.E.; Ross, R.D.; Tilley, J.M.; Vargo-Gogola, T.; Roeder, R.K. Gold nanoparticles as contrast agents in x-ray imaging and computed tomography. Nanomedicine 2015, 10, 321–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Ross, R.D.; Roeder, R.K. Preparation of functionalized gold nanoparticles as a targeted X-ray contrast agent for damaged bone tissue. Nanoscale 2010, 2, 582–586. [Google Scholar] [CrossRef]
- Caro, C.; Dalmases, M.; Figuerola, A.; García-Martín, M.L.; Leal, M.P. Highly water-stable rare ternary Ag–Au–Se nanocomposites as long blood circulation time X-ray computed tomography contrast agents. Nanoscale 2017, 9, 7242–7251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caro, C.; Gámez, F.; Quaresma, P.; Páez-Muñoz, J.M.; Domínguez, A.; Pearson, J.R.; Pernía Leal, M.; Beltrán, A.M.; Fernandez-Afonso, Y.; De la Fuente, J.M.; et al. Fe(3)O(4)-Au core-shell nanoparticles as a multimodal platform for in vivo imaging and focused photothermal therapy. Pharmaceutics 2021, 13, 416. [Google Scholar] [CrossRef] [PubMed]
- Regulla, D.F.; Hieber, L.B.; Seidenbusch, M. Physical and biological interface dose effects in tissue due to x-ray-induced release of secondary radiation from metallic gold surfaces. Radiat. Res. 1998, 150, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Regulla, D.; Schmid, E.; Friedland, W.; Panzer, W.; Heinzmann, U.; Harder, D. Enhanced values of the RBE and H ratio for cytogenetic effects induced by secondary electrons from an X-irradiated gold gurface. Radiat. Res. 2002, 158, 505–515. [Google Scholar] [CrossRef]
- Hainfeld, J.F.; Slatkin, D.N.; Smilowitz, H.M. The use of gold nanoparticles to enhance radiotherapy in mice. Phys. Med. Biol. 2004, 49, N309–N315. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H. Estimation of tumour dose enhancement due to gold nanoparticles during typical radiation treatments: A preliminary Monte Carlo study. Phys. Med. Biol. 2005, 50, N163–N173. [Google Scholar] [CrossRef] [PubMed]
- Bergs, J.W.; Wacker, M.G.; Hehlgans, S.; Piiper, A.; Multhoff, G.; Rödel, C.; Rödel, F. The role of recent nanotechnology in enhancing the efficacy of radiation therapy. Biochim. Biophys. Acta 2015, 1856, 130–143. [Google Scholar] [CrossRef]
- Cooper, D.R.; Bekah, D.; Nadeau, J.L. Gold nanoparticles and their alternatives for radiation therapy enhancement. Front. Chem. 2014, 2, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zygmanski, P.; Sajo, E. Nanoscale radiation transport and clinical beam modeling for gold nanoparticle dose enhanced radiotherapy (GNPT) using X-rays. Br. J. Radiol. 2016, 89, 20150200. [Google Scholar] [CrossRef] [Green Version]
- Ngwa, W.; Kumar, R.; Sridhar, S.; Korideck, H.; Zygmanski, P.; Cormack, R.A.; Berbeco, R.; Makrigiorgos, G.M. Targeted radiotherapy with gold nanoparticles: Current status and future perspectives. Nanomedicine 2014, 9, 1063–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, S.J.; Mendenhall, M.H.; Jain, S.; Currell, F. Radiotherapy in the presence of contrast agents: A general figure of merit and its application to gold nanoparticles. Phys. Med. Biol. 2008, 53, 5635–5651. [Google Scholar] [CrossRef]
- Jones, B.L.; Krishnan, S.; Cho, S.H. Estimation of microscopic dose enhancement factor around gold nanoparticles by Monte Carlo calculations. Med. Phys. 2010, 37, 3809–3816. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.K.; Chow, J.C.; Chithrani, B.D.; Lee, M.J.; Oms, B.; Jaffray, D.A. Irradiation of gold nanoparticles by x-rays: Monte Carlo simulation of dose enhancements and the spatial properties of the secondary electrons production. Med. Phys. 2011, 38, 624–631. [Google Scholar] [CrossRef]
- Lechtman, E.; Mashouf, S.; Chattopadhyay, N.; Keller, B.M.; Lai, P.; Cai, Z.; Reilly, R.M.; Pignol, J.P. A Monte Carlo based model of gold nanoparticle radiosensitization accounting for increased radiobiological effectiveness. Phys. Med. Biol. 2013, 58, 3075–3087. [Google Scholar] [CrossRef]
- Douglass, M.; Bezak, E.; Penfold, S. Monte Carlo investigation of the increased radiation deposition due to gold nanoparticles using kilovoltage and megavoltage photons in a 3D randomized cell model. Med. Phys. 2013, 40, 071710. [Google Scholar] [CrossRef]
- Zygmanski, P.; Liu, B.; Tsiamas, P.; Cifter, F.; Petersheim, M.; Hesser, J.; Sajo, E. Dependence of Monte Carlo microdosimetric computations on the simulation geometry of gold nanoparticles. Phys. Med. Biol. 2013, 58, 7961–7977. [Google Scholar] [CrossRef] [PubMed]
- Li, W.B.; Müllner, M.; Greiter, M.B.; Bissardon, C.; Xie, W.Z.; Schlattl, H.; Oeh, U.; Li, J.L.; Hoeschen, C. Monte Carlo simulations of dose enhancement around gold nanoparticles used as x-ray imaging contrast agents and radiosensitizers. Proc. SPIE 2014, 9033, 90331K. [Google Scholar]
- Xie, W.Z.; Friedland, W.; Li, W.B.; Li, C.Y.; Oeh, U.; Qiu, R.; Li, J.L.; Hoeschen, C. Simulation on the molecular radiosensitization effect of gold nanoparticles in cells irradiated by x-rays. Phys. Med. Biol. 2015, 60, 6195–6212. [Google Scholar] [CrossRef]
- Lin, Y.; McMahon, S.J.; Scarpelli, M.; Paganetti, H.; Schuemann, J. Comparing gold nano-particle enhanced radiotherapy with protons, megavoltage photons and kilovoltage photons: A Monte Carlo simulation. Phys. Med. Biol. 2014, 59, 7675–7689. [Google Scholar] [CrossRef] [PubMed]
- Vlastou, E.; Diamantopoulos, S.; Efstathopoulos, E.P. Monte Carlo studies in Gold Nanoparticles enhanced radiotherapy: The impact of modelled parameters in dose enhancement. Phys. Med. 2020, 80, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Sajo, E.; Zygmanski, P. Nanoparticle Enhanced Radiation Therapy. Principles, Methods and Applications; IOP Publishing: Bristol, UK, 2020. [Google Scholar] [CrossRef]
- Li, W.B.; Belchior, A.; Beuve, M.; Chen, Y.Z.; Di Maria, S.; Friedland, W.; Gervais, B.; Heide, B.; Hocine, N.; Ipatov, A.; et al. Intercomparison of dose enhancement ratio and secondary electron spectra for gold nanoparticles irradiated by X-rays calculated using multiple Monte Carlo simulation codes. Phys. Med. 2020, 69, 147–163. [Google Scholar] [CrossRef] [Green Version]
- Fink, S.L.; Cookson, B.T. Apoptosis, pyroptosis, and necrosis: Mechanistic description of dead and dying eukaryotic cells. Infect. Immun. 2005, 73, 1907–1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudek, B.; McNamara, A.; Ramos-Méndez, J.; Byrne, H.; Kuncic, Z.; Schuemann, J. Radio-enhancement by gold nanoparticles and their impact on water radiolysis for x-ray, proton and carbon-ion beams. Phys. Med. Biol. 2019, 64, 175005. [Google Scholar] [CrossRef]
- Zhang, Z.; Berg, A.; Levanon, H.; Fessenden, R.W.; Meisel, D. On the interactions of free radicals with gold nanoparticles. J. Am. Chem. Soc. 2003, 125, 7959–7963. [Google Scholar] [CrossRef]
- Poignant, F.; Charfi, H.; Chan, C.-H.; Dumont, E.; Loffreda, D.; Gervais, B.; Beuve, M. Monte Carlo simulation of free radical production under keV photon irradiation of gold nanoparticle aqueous solution. Part II: Local primary chemical boost. Radiat. Phys. Chem. 2021, 179, 109161. [Google Scholar] [CrossRef]
- Poignant, F.; Charfi, H.; Chan, C.-H.; Dumont, E.; Loffreda, D.; Testa, É.; Gervais, B.; Beuve, M. Monte Carlo simulation of free radical production under keV photon irradiation of gold nanoparticle aqueous solution. Part I: Global primary chemical boost. Radiat. Phys. Chem. 2020, 172, 108790. [Google Scholar] [CrossRef]
- Schuemann, J.; Berbeco, R.; Chithrani, D.B.; Cho, S.H.; Kumar, R.; McMahon, S.J.; Sridhar, S.; Krishnan, S. Roadmap to clinical use of gold nanoparticles for radiation sensitization. Int. J. Radiat. Oncol. Biol. Phys. 2016, 94, 189–205. [Google Scholar] [CrossRef] [Green Version]
- Schuemann, J.; Bagley, A.F.; Berbeco, R.; Bromma, K.; Butterworth, K.T.; Byrne, H.L.; Chithrani, B.D.; Cho, S.H.; Cook, J.R.; Favaudon, V.; et al. Roadmap for metal nanoparticles in radiation therapy: Current status, translational challenges, and future directions. Phys. Med. Biol. 2020, 65, 21rm02. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Stewart, J.; Allen, C.; Jaffray, D.A. Intracellular uptake, transport, and processing of nanostructures in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2009, 5, 118–127. [Google Scholar] [CrossRef]
- Chithrani, D.B.; Chan, C.W.W. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett 2007, 7, 1542–1550. [Google Scholar] [CrossRef]
- Chithrani, D.B.; Dunne, M.; Stewart, J.; Allen, C.; Jaffray, D.A. Cellular uptake and transport of gold nanoparticles incorporated in a liposomal carrier. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, D.B.; Ghazani, A.A.; Chan, C.W.W. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef]
- Chithrani, D.B.; Jelveh, S.; Jalali, F.; van Prooijen, M.; Allen, C.; Bristow, R.G.; Hill, R.P.; Jaffray, D.A. Gold nanoparticles as radiation sensitizers in cancer therapy. Radiat. Res. 2010, 173, 719–728. [Google Scholar] [CrossRef]
- Yang, C.; Uertz, J.; Yohan, D.; Chithrani, B.D. Peptide modified gold nanoparticles for improved cellular uptake, nuclear transport, and intracellular retention. Nanoscale 2014, 6, 12026–12033. [Google Scholar] [CrossRef]
- Chattopadhyay, N.; Cai, Z.; Kwon, Y.L.; Lechtman, E.; Pignol, J.-P.; Reilly, R.M. Molecularly targeted gold nanoparticles enhance the radiation response of breast cancer cells and tumor xenografts to X-radiation. Breast Cancer Res. Treat. 2013, 137, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, N.; Cai, Z.; Pignol, J.P.; Keller, B.; Lechtman, E.; Bendayan, R.; Reilly, R.M. Design and characterization of HER-2-targeted gold nanoparticles for enhanced X-radiation treatment of locally advanced breast cancer. Mol. Pharm. 2010, 7, 2194–2206. [Google Scholar] [CrossRef] [PubMed]
- Mueller, R.; Yasmin-Karim, S.; DeCosmo, K.; Vazquez-Pagan, A.; Sridhar, S.; Kozono, D.; Hesser, J.; Ngwa, W. Increased carcinoembryonic antigen expression on the surface of lung cancer cells using gold nanoparticles during radiotherapy. Phys. Med. 2020, 76, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Yu, B.; Jin, X.; Zhao, T.; Ye, F.; Liu, X.; Li, P.; Zheng, X.; Chen, W.; Li, Q. Therapeutic efficacy of carbon ion irradiation enhanced by 11-MUA-capped gold nanoparticles: An in vitro and in vivo study. Int. J. Nanomed. 2021, 16, 4661–4674. [Google Scholar] [CrossRef]
- Berbeco, R.I.; Korideck, H.; Ngwa, W.; Kumar, R.; Patel, J.; Sridhar, S.; Johnson, S.; Price, B.D.; Kimmelman, A.; Makrigiorgos, G.M. DNA damage enhancement from gold nanoparticles for clinical MV photon beams. Radiat. Res. 2012, 178, 604–608. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Coulter, J.A.; Hounsell, A.R.; Butterworth, K.T.; McMahon, S.J.; Hyland, W.B.; Muir, M.F.; Dickson, G.R.; Prise, K.M.; Currell, F.J.; et al. Cell-specic radiosensitization by gold nanoparticles at megavoltage radiation energies. Int. J. Radiat. Oncol. Biol. Phys. 2011, 79, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-K.; Seo, S.-J.; Kim, H.-T.; Kim, K.-H.; Chung, M.-H.; Kim, K.-R.; Ye, S.-J. Enhanced proton treatment in mouse tumors through proton irradiated nanoradiator effects on metallic nanoparticles. Phys. Med. Biol. 2012, 57, 8309–8323. [Google Scholar] [CrossRef]
- Kaur, H.; Pujari, G.; Semwal, M.K.; Sarma, A.; Avasthi, D.K. In vitro studies on radiosensitization effect of glucose capped gold nanoparticles in photon and ion irradiation of HeLa cells. Nucl. Instr. Meth. Phys. Res. B 2013, 301, 7–11. [Google Scholar] [CrossRef]
- Cho, E.C.; Zhang, Q.; Xia, Y. The effect of sedimentation and diffusion on cellular uptake of gold nanoparticles. Nat. Nanotechnol. 2011, 6, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Taylor, U.; Barchanski, A.; GarreIs, W.; Klein, S.; Kues, W.; Barcikowski, S.; Rath, D. Chapter 12. Toxicity of gold nanoparticles on somatic and reproductive cells. In Nano-Biotechnology for Biomedical and Diagnostic Research; Zahavy, E., Ordentlich, A., Yitzhaki, S., Shafferman, A., Eds.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Sherman, M.; Multhoff, G. Heat shock proteins in cancer. Ann. N. Y. Acad. Sci. 2007, 1113, 192–201. [Google Scholar] [CrossRef]
- Stangl, S.; Gehrmann, M.; Riegger, J.; Kuhs, I.; Riederer, I.; Sievert, W.; Hube, K.; Mocikat, R.; Dressel, R.; Kremmer, E.; et al. Targeting membrane heat-shock protein 70 (Hsp70) on tumors by cmHsp70.1 antibody. Proc. Natl. Acad. Sci. USA 2011, 108, 733–738. [Google Scholar] [CrossRef] [Green Version]
- Gehrmann, M.; Radons, J.; Molls, M.; Multhoff, G. The therapeutic implications of clinically applied modifiers of heat shock protein 70 (Hsp70) expression by tumor cells. Cell Stress Chaperones 2008, 13, 1–10. [Google Scholar] [CrossRef]
- Stangl, S.; Gehrmann, M.; Dressel, R.; Alves, F.; Dullin, C.; Themelis, G.; Ntziachristos, V.; Staeblein, E.; Walch, A.; Winkelmann, I.; et al. In vivo imaging of CT26 mouse tumours by using cmHsp70.1 monoclonal antibody. J. Cell Mol. Med. 2011, 15, 874–887. [Google Scholar] [CrossRef] [Green Version]
- Gehrmann, M.K.; Kimm, M.A.; Stangl, S.; Schmid, T.E.; Noël, P.B.; Rummeny, E.J.; Multhoff, G. Imaging of Hsp70-positive tumors with cmHsp70.1 antibody-conjugated gold nanoparticles. Int. J. Nanomed. 2015, 10, 5687–5700. [Google Scholar] [CrossRef] [Green Version]
- Shevtsov, M.A.; Nikolaev, B.P.; Ryzhov, V.A.; Yakovleva, L.Y.; Marchenko, Y.Y.; Parr, M.A.; Rolich, V.I.; Mikhrina, A.L.; Dobrodumov, A.V.; Pitkini, E.; et al. Ionizing radiation improves glioma-specific targeting of superparamagnetic iron oxide nanoparticles conjugated with cmHsp70.1 monoclonal antibodies (SPION-cmHsp70.1). Nanoscale 2015, 7, 20652. [Google Scholar] [CrossRef] [PubMed]
- Hainfeld, J.F.; Slatkin, D.N.; Focella, T.M.; Smilowitz, H.M. Gold nanoparticles: A new X-ray contrast agent. Br. J. Radiol. 2006, 79, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Hainfeld, J.F.; Smilowitz, H.M.; O’Connor, M.J.; Dilmanian, F.A.; Slatkin, D.N. Gold nanoparticle imaging and radiotherapy of brain tumors in mice. Nanomedicine 2013, 8, 1601–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popovtzer, R.; Agrawal, A.; Kotov, N.A.; Popovtzer, A.; Balter, J.; Carey, T.E.; Kopelman, R. Targeted gold nanoparticles enable molecular CT imaging of cancer. Nano Lett. 2008, 8, 4593–4596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazalova, M.; Kuang, Y.; Pratx, G.; Xing, L. Investigation of X-ray fluorescence computed tomography (XFCT) and K-edge imaging. IEEE Trans. Med Imaging 2012, 31, 1620–1627. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Hoeschen, C.; Grüner, F.; Johnson, T.R.C. X-ray fluorescence molecular imaging of high-Z tracers: Investigation of a novel analyzer based setup. SPIE 2014, 9033, 90331J. [Google Scholar]
- Manohar, N.; Reynoso, F.; Jayarathna, S.; Moktan, H.; Ahmed, M.F.; Diagaradjane, P.; Krishnan, S.; Cho, S.H. High-sensitivity imaging and quantification of intratumoral distributions of gold nanoparticles using a benchtop x-ray fluorescence imaging system. Opt. Lett. 2019, 44, 5314–5317. [Google Scholar] [CrossRef] [PubMed]
- Manohar, N.; Reynoso, F.J.; Diagaradjane, P.; Krishnan, S.; Cho, S.H. Quantitative imaging of gold nanoparticle distribution in a tumor-bearing mouse using benchtop x-ray fluorescence computed tomography. Sci. Rep. 2016, 6, 22079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manohar, N.; Jones, B.L.; Cho, S.H. Improving x-ray fluorescence signal for benchtop polychromatic cone-beam x-ray fluorescence computed tomography by incident x-ray spectrum optimization: A Monte Carlo study. Med. Phys. 2014, 41, 101906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manohar, N.; Reynoso, F.J.; Cho, S.H. Technical note: A benchtop cone-beam x-ray fluorescence computed tomography (XFCT) system with a high-power x-ray source and transmission CT imaging capability. Med. Phys. 2018, 45, 4652–4659. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.L.; Manohar, N.; Reynoso, F.; Karellas, A.; Cho, S.H. Experimental demonstration of benchtop x-ray fluorescence computed tomography (XFCT) of gold nanoparticle-loaded objects using lead- and tin-filtered polychromatic cone-beams. Phys. Med. Biol. 2012, 57, N457–N467. [Google Scholar] [CrossRef] [PubMed]
- Kimm, M.A.; Shevtsov, M.; Werner, C.; Sievert, W.; Zhiyuan, W.; Schoppe, O.; Menze, B.H.; Rummeny, E.J.; Proksa, R.; Bystrova, O.; et al. Gold nanoparticle mediated multi-modal CT imaging of Hsp70 membrane-positive tumors. Cancers 2020, 12, 1331. [Google Scholar] [CrossRef]
- Abed, Z.; Beik, J.; Laurent, S.; Eslahi, N.; Khani, T.; Davani, E.S.; Ghaznavi, H.; Shakeri-Zadeh, A. Iron oxide-gold core-shell nano-theranostic for magnetically targeted photothermal therapy under magnetic resonance imaging guidance. J. Cancer Res. Clin. Oncol. 2019, 145, 1213–1219. [Google Scholar] [CrossRef]
- Beik, J.; Asadi, M.; Khoei, S.; Laurent, S.; Abed, Z.; Mirrahimi, M.; Farashahi, A.; Hashemian, R.; Ghaznavi, H.; Shakeri-Zadeh, A. Simulation-guided photothermal therapy using MRI-traceable iron oxide-gold nanoparticle. J. Photochem. Photobiol. B 2019, 199, 111599. [Google Scholar] [CrossRef] [PubMed]
- Shevtsov, M.; Multhoff, G. Recent developments of magnetic nanoparticles for theranostics of brain tumor. Curr. Drug Metab. 2016, 17, 737–744. [Google Scholar] [CrossRef]
- Shevtsov, M.; Stangl, S.; Nikolaev, B.; Yakovleva, L.; Marchenko, Y.; Tagaeva, R.; Sievert, W.; Pitkin, E.; Mazur, A.; Tolstoy, P.; et al. Granzyme B functionalized nanoparticles targeting membrane Hsp70-positive tumors for multimodal cancer theranostics. Small 2019, 15, e1900205. [Google Scholar] [CrossRef]
- Asadi, M.; Beik, J.; Hashemian, R.; Laurent, S.; Farashahi, A.; Mobini, M.; Ghaznavi, H.; Shakeri-Zadeh, A. MRI-based numerical modeling strategy for simulation and treatment planning of nanoparticle-assisted photothermal therapy. Phys. Med. 2019, 66, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Klapproth, A.P.; Shevtsov, M.; Stangl, S.; Li, W.B.; Multhoff, G. A new pharmacokinetic model describing the biodistribution of intravenously and intratumorally administered superparamagnetic iron oxide nanoparticles (SPIONs) in a GL261 xenograft glioblastoma model. Int. J. Nanomed. 2020, 15, 4677–4689. [Google Scholar] [CrossRef] [PubMed]
- Schlomka, J.P.; Roessl, E.; Dorscheid, R.; Dill, S.; Martens, G.; Istel, T.; Bäumer, C.; Herrmann, C.; Steadman, R.; Zeitler, G.; et al. Experimental feasibility of multi-energy photon-counting K-edge imaging in pre-clinical computed tomography. Phys. Med. Biol. 2008, 53, 4031–4047. [Google Scholar] [CrossRef] [PubMed]
- Poludniowski, G.; Landry, G.; DeBlois, F.; Evans, P.M.; Verhaegen, F. SpekCalc: A program to calculate photon spectra from tungsten anode x-ray tubes. Phys. Med. Biol. 2009, 54, N433–N438. [Google Scholar] [CrossRef] [Green Version]
- Poludniowski, G.G. Calculation of x-ray spectra emerging from an x-ray tube. Part II. X-ray production and filtration in x-ray targets. Med Phys. 2007, 34, 2175–2186. [Google Scholar] [CrossRef] [PubMed]
- Salvat, F. PENELOPE-2018. A Code System for Monte Carlo Simulation of Electron and Photon Transport; NEA/MBDAV/R(2019)1; Nuclear Energy Agency/Organisation for Economic Co-Operatrion and Developement: Paris, France, 2019. [Google Scholar]
- Incerti, S.; Baldacchino, G.; Bernal, M.; Capra, R.; Champion, C.; Francis, Z.; Guatelli, S.; Guèye, P.; Mantero, A.; Mascialino, B.; et al. The Geant4-DNA project. Int. J. Model Simul. Sci. Comput. 2010, 1, 157–178. [Google Scholar] [CrossRef]
- Li, J.; Li, C.; Qiu, R.; Yan, C.; Xie, W.; Wu, Z.; Zeng, Z.; Tung, C. DNA strand breaks induced by electrons simulated with Nanodosimetry Monte Carlo Simulation Code: NASIC. Radiat. Prot. Dosim. 2015, 166, 38–43. [Google Scholar] [CrossRef]
- Hill, R.; Healy, B.; Holloway, L.; Kuncic, Z.; Thwaites, D.; Baldock, C. Advances in kilovoltage x-ray beam dosimetry. Phys. Med. Biol. 2014, 59, R183–R231. [Google Scholar] [CrossRef]
- Friedland, W.; Dingfelder, M.; Kundrát, P.; Jacob, P. Track structures, DNA targets and radiation effects in the biophysical Monte Carlo simulation code PARTRAC. Mutat. Res. 2011, 711, 28–40. [Google Scholar] [CrossRef]
- Schuemann, J.; McNamara, A.L.; Ramos-Mendez, J.; Perl, J.; Held, K.D.; Paganetti, H.; Incerti, S.; Faddegon, B. TOPAS-nBio: An extension to the TOPAS simulation toolkit for cellular and sub-cellular radiobiology. Radiat. Res. 2019, 191, 125–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perl, J.; Shin, J.; Schumann, J.; Faddegon, B.; Paganetti, H. TOPAS: An innovative proton Monte Carlo platform for research and clinical applications. Med. Phys. 2012, 39, 6818–6837. [Google Scholar] [CrossRef]
- Ramos-Méndez, J.; Perl, J.; Schuemann, J.; McNamara, A.; Paganetti, H.; Faddegon, B. Monte Carlo simulation of chemistry following radiolysis with TOPAS-nBio. Phys. Med. Biol. 2018, 63, 105014. [Google Scholar] [CrossRef]
- Karamitros, M.; Luan, S.; Bernal, M.A.; Allison, J.; Baldacchino, G.; Davidkova, M.; Francis, Z.; Friedland, W.; Ivantchenko, V.; Ivantchenko, A.; et al. Diffusion-controlled reactions modeling in Geant4-DNA. J. Comput. Phys. 2014, 274, 841–882. [Google Scholar] [CrossRef]
- Zhu, H.; McNamara, A.L.; Ramos-Mendez, J.; McMahon, S.J.; Henthorn, N.T.; Faddegon, B.; Held, K.D.; Perl, J.; Li, J.; Paganetti, H.; et al. A parameter sensitivity study for simulating DNA damage after proton irradiation using TOPAS-nBio. Phys. Med. Biol. 2020, 65, 085015. [Google Scholar] [CrossRef] [PubMed]
- Klapproth, A.P.; Schuemann, J.; Stangl, S.; Xie, T.; Li, W.B.; Multhoff, G. Multi-scale Monte Carlo simulations of gold nanoparticle induced DNA damages for kilovoltage X-ray irradiation in a xenograft mouse model using TOPAS-nBio. Cancer Nanotechnol. 2021, 12, 644–653. [Google Scholar] [CrossRef]
- Xie, T.; Zaidi, H. Monte Carlo-based evaluation of S-values in mouse models for positron-emitting radionuclides. Phys. Med. Biol. 2013, 58, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.; Armour, E.; Kazanzides, P.; Iordachita, I.; Tryggestad, E.; Deng, H.; Matinfar, M.; Kennedy, C.; Liu, Z.; Chan, T.; et al. High-resolution, small animal radiation research platform with x-ray tomographic guidance capabilities. Int. J. Radiat. Oncol. Biol. Phys. 2008, 71, 1591–1599. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Chen, Y.; Sung, W.; McNamara, A.L.; Tran, L.T.; Burigo, L.N.; Rosenfeld, A.B.; Li, J.; Faddegon, B.; Schuemann, J.; et al. The microdosimetric extension in TOPAS: Development and comparison with published data. Phys. Med. Biol. 2019, 64, 145004. [Google Scholar] [CrossRef]
- Lampe, N.; Karamitros, M.; Breton, V.; Brown, J.M.C.; Kyriakou, I.; Sakata, D.; Sarramia, D.; Incerti, S. Mechanistic DNA damage simulations in Geant4-DNA part 1: A parameter study in a simplified geometry. Phys. Med. 2018, 48, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Meylan, S.; Incerti, S.; Karamitros, M.; Tang, N.; Bueno, M.; Clairand, I.; Villagrasa, C. Simulation of early DNA damage after the irradiation of a fibroblast cell nucleus using Geant4-DNA. Sci. Rep. 2017, 7, 11923. [Google Scholar] [CrossRef] [PubMed]
- Sakata, D.; Lampe, N.; Karamitros, M.; Kyriakou, I.; Belov, O.; Bernal, M.A.; Bolst, D.; Bordage, M.C.; Breton, V.; Brown, J.M.C.; et al. Evaluation of early radiation DNA damage in a fractal cell nucleus model using Geant4-DNA. Phys. Med. 2019, 62, 152–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, M.A.; de Almeida, C.E.; Sampaio, C.; Incerti, S.; Champion, C.; Nieminen, P. The invariance of the total direct DNA strand break yield. Med. Phys. 2011, 38, 4147–4153. [Google Scholar] [CrossRef] [PubMed]
- Tartier, L.; Michalik, V.; Spotheim-Maurizot, M.; Rahmouni, A.R.; Sabattier, R.; Charlier, M. Radiolytic signature of Z-DNA. Nucleic Acids Res. 1994, 22, 5565–5570. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, E.; Rezin, G.T.; Zanoni, E.T.; de Souza Notoya, F.; Leffa, D.D.; Damiani, A.P.; Daumann, F.; Rodriguez, J.C.O.; Benavides, R.; da Silva, L.; et al. Acute and chronic administration of gold nanoparticles cause DNA damage in the cerebral cortex of adult rats. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2014, 766–767, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Chaves, C.; Soto-Alvaredo, J.; Montes-Bayon, M.; Bettmer, J.; Llopis, J.; Sanchez-Gonzalez, C. Gold nanoparticles: Distribution, bioaccumulation and toxicity. In vitro and in vivo studies. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1–12. [Google Scholar] [CrossRef]
- May, S.; Hirsch, C.; Rippl, A.; Bohmer, N.; Kaiser, J.P.; Diener, L.; Wichser, A.; Bürkle, A.; Wick, P. Transient DNA damage following exposure to gold nanoparticles. Nanoscale 2018, 10, 15723–15735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotiropoulos, M.; Henthorn, N.T.; Warmenhoven, J.W.; Mackay, R.I.; Kirkby, K.J.; Merchant, M.J. Modelling direct DNA damage for gold nanoparticle enhanced proton therapy. Nanoscale 2017, 9, 18413–18422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, H.; Shi, L.; Xu, J.; Duan, G.; Yang, H. A focus on the genotoxicity of gold nanoparticles. Nanomedicine 2020, 15, 319–323. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Huang, C.; Chen, X.; Yi, Z.; Sanche, L. Chemical radiosensitivity of DNA induced by gold nanoparticles. J. Biomed. Nanotechnol. 2015, 11, 478–485. [Google Scholar] [CrossRef]
- Kirkby, C.; Ghasroddashti, E. Targeting mitochondria in cancer cells using gold nanoparticle-enhanced radiotherapy: A Monte Carlo study. Med. Phys. 2015, 42, 1119–1128. [Google Scholar] [CrossRef]
- Taggart, L.E.; McMahon, S.J.; Currell, F.J.; Prise, K.M.; Butterworth, K.T. The role of mitochondrial function in gold nanoparticle mediated radiosensitisation. Cancer Nanotechnol. 2014, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Scholz, M.; Kraft, G. Calculation of heavy ion inactivation probabilities based on track structure, X ray sensitivity and target size. Radiat. Prot. Dosim. 1994, 52, 29–33. [Google Scholar] [CrossRef]
- Brown, J.M.C.; Currell, F.J. A local effect model-based interpolation framework for experimental nanoparticle radiosensitisation data. Cancer Nanotechnol. 2017, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Sung, W.; Ye, S.J. Microdosimetric-kinetic model for radio-enhancement of gold nanoparticles: Comparison with LEM. Radiat. Res. 2021, 195, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Melo-Bernal, W.; Chernov, G.; Barboza-Flores, M.; Chernov, V. Quantification of the radiosensitization effect of high-Znanoparticles on photon irradiated cells: Combining Monte Carlo simulations and an analytical approach to the local effect model. Phys. Med. Biol. 2021, 66, 135007. [Google Scholar] [CrossRef]
- Kellerer, A.M.; Rossi, H.H. RBE and the primary mechanism of radiation action. Radiat. Res. 1971, 47, 15–34. [Google Scholar] [CrossRef] [Green Version]
- Chadwick, K.H.; Leenhouts, H.P. A molecular theory of cell survival. Phys. Med. Biol. 1973, 18, 78–87. [Google Scholar] [CrossRef]
- Wirthl, B.; Kremheller, J.; Schrefler, B.A.; Wall, W.A. Extension of a multiphase tumour growth model to study nanoparticle delivery to solid tumours. PLoS ONE 2020, 15, e0228443. [Google Scholar] [CrossRef] [Green Version]
- Rabus, H.; Gargioni, E.; Li, W.B.; Nettelbeck, H.; Villagrasa, C. Determining dose enhancement factors of high-Z nanoparticles from simulations where lateral secondary particle disequilibrium exists. Phys. Med. Biol. 2019, 64, 155016. [Google Scholar] [CrossRef] [Green Version]
- Sakata, D.; Kyriakou, I.; Okada, S.; Tran, H.N.; Lampe, N.; Guatelli, S.; Bordage, M.-C.; Ivanchenko, V.; Murakami, K.; Sasaki, T.; et al. Geant4-DNA track-structure simulations for gold nanoparticles: The importance of electron discrete models in nanometer volumes. Med. Phys. 2018, 45, 2230–2242. [Google Scholar] [CrossRef] [Green Version]
- Stumpf, V.; Kolorenc, P.; Gokhberg, K.; Cederbaum, L.S. Efficient pathway to neutralization of multiply charged ions produced in Auger processes. Phys. Rev. Lett. 2013, 110, 258302. [Google Scholar] [CrossRef] [PubMed]
- Kirkby, C.; Koger, B. Nanoparticle-aided radiation therapy: Challenges of treatment planning. In Nanoparticle Enhanced Radiation Therapy; IOP Publishing: Bristol, UK, 2020; pp. 6-1–6-31. [Google Scholar] [CrossRef]
- Kuncic, Z.; Lacombe, S. Nanoparticle radio-enhancement: Principles, progress and application to cancer treatment. Phys. Med. Biol. 2018, 63, 02tr01. [Google Scholar] [CrossRef]
- Villagomez-Bernabe, B.; Currell, F.J. Physical radiation enhancement effects around clinically relevant clusters of nanoagents in biological systems. Sci. Rep. 2019, 9, 8156. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chun, S.H.; Amornkitbamrung, L.; Song, C.; Yuk, J.S.; Ahn, S.Y.; Kim, B.W.; Lim, Y.T.; Oh, B.-K.; Um, S.H. Gold nanoparticle clusters for the investigation of therapeutic efficiency against prostate cancer under near-infrared irradiation. Nano Converg. 2020, 7, 5. [Google Scholar] [CrossRef] [PubMed]




















| Parameter | Unit | Values |
|---|---|---|
| Peak voltage | kVp | 50, 60, 80, 100, 150 and 200 |
| Energy bin | keV | 0.5 |
| Angle theta | degree | 20 |
| Air thickness | mm | 470 |
| Beryllium thickness | mm | 0.8 |
| Aluminum thickness | mm | 3.9 |
| Nf | 0.68 | |
| P | 0.33 |
| DSB Class | Q | p | R2 | |||
|---|---|---|---|---|---|---|
| k1 (10−3) | k2 (10−5) | Notation | Value (10−3) | |||
| I | 1.090 | 5.917 | −1.879 | p | 9.944 | 0.9893 |
| II | 1.037 | 7.275 | −2.296 | 3.088 | 0.9987 | |
| 30.68 | ||||||
| Systemic Circulation | Transfer Rate to Blood (h−1) | Clearance Rate from Blood (h−1) |
|---|---|---|
| Tumor | 327.4 | 40.06 |
| Liver | 0.1159 | 5.067 × 10−4 |
| Spleen | 9.78 × 10−4 | 1.409 × 10−5 |
| Kidneys | 3.804 | 15.66 |
| Colon | 0.7151 | 689.6 |
| Stomach | 7.411 | 896.0 |
| Lungs | 1.903 | 148.5 |
| Small intestine | 68.93 | 59.41 |
| Muscle | 194.2 | 62.39 |
| Bone | 62.6 | 38.58 |
| Skin | 9.38 | 27.58 |
| Brain | 5.439 | 795.6 |
| Heart | 2.416 | 490.5 |
| Alimentary tract | ||
| Donor | Receptor | Transfer rate (h−1) |
| Liver | Small intestine | 4.947 × 10−4 |
| Stomach | Small intestine | 64.71 |
| Small intestine | Colon | 3.447 |
| Colon | Excretion | 4.244 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.B.; Stangl, S.; Klapproth, A.; Shevtsov, M.; Hernandez, A.; Kimm, M.A.; Schuemann, J.; Qiu, R.; Michalke, B.; Bernal, M.A.; et al. Application of High-Z Gold Nanoparticles in Targeted Cancer Radiotherapy—Pharmacokinetic Modeling, Monte Carlo Simulation and Radiobiological Effect Modeling. Cancers 2021, 13, 5370. https://doi.org/10.3390/cancers13215370
Li WB, Stangl S, Klapproth A, Shevtsov M, Hernandez A, Kimm MA, Schuemann J, Qiu R, Michalke B, Bernal MA, et al. Application of High-Z Gold Nanoparticles in Targeted Cancer Radiotherapy—Pharmacokinetic Modeling, Monte Carlo Simulation and Radiobiological Effect Modeling. Cancers. 2021; 13(21):5370. https://doi.org/10.3390/cancers13215370
Chicago/Turabian StyleLi, Wei Bo, Stefan Stangl, Alexander Klapproth, Maxim Shevtsov, Alicia Hernandez, Melanie A. Kimm, Jan Schuemann, Rui Qiu, Bernhard Michalke, Mario A. Bernal, and et al. 2021. "Application of High-Z Gold Nanoparticles in Targeted Cancer Radiotherapy—Pharmacokinetic Modeling, Monte Carlo Simulation and Radiobiological Effect Modeling" Cancers 13, no. 21: 5370. https://doi.org/10.3390/cancers13215370
APA StyleLi, W. B., Stangl, S., Klapproth, A., Shevtsov, M., Hernandez, A., Kimm, M. A., Schuemann, J., Qiu, R., Michalke, B., Bernal, M. A., Li, J., Hürkamp, K., Zhang, Y., & Multhoff, G. (2021). Application of High-Z Gold Nanoparticles in Targeted Cancer Radiotherapy—Pharmacokinetic Modeling, Monte Carlo Simulation and Radiobiological Effect Modeling. Cancers, 13(21), 5370. https://doi.org/10.3390/cancers13215370