SILAC-Based Quantitative Proteomic Analysis of Oxaliplatin-Resistant Pancreatic Cancer Cells
Abstract
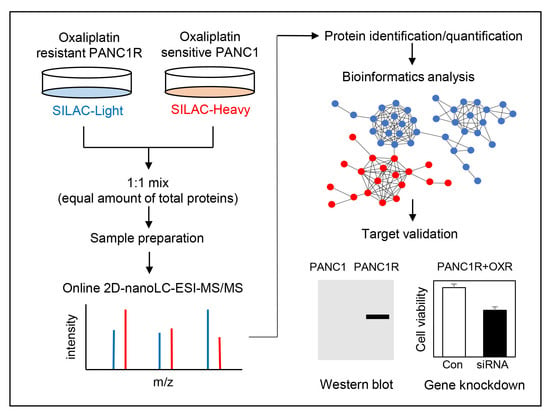
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. The Establishment and Validation of Oxaliplatin-Resistant PANC-1 Cells
2.2. Quantitative Proteomic Analysis of Oxaliplatin-Resistant and Sensitive PANC-1 Cells
2.3. Verification of Differentially Expressed Proteins between Oxaliplatin Sensitive and Resistant PANC-1 Cells by Western Blot
2.4. MARCKS or WLS Was a Significant Factor for Chemoresistant in PANC-1R Cells
2.5. Inhibition of MARCKS and WLS Increased Oxaliplatin-Mediated Cell Death in Chemoresistant PANC-1R Cells
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Statistical Rationale
4.2. Establishment of an Oxaliplatin-Resistant Pancreatic Cancer Cell Line
4.3. Cell Viability Assay
4.4. Colony Forming Assay
4.5. Stable Isotope Labelling with Amino Acids in Cell Culture (SILAC)
4.6. Sample Preparation for Proteomic Analysis
4.7. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
4.8. Data Analysis
4.9. Western Blot Analysis
4.10. RNA Isolation and qRT-PCR
4.11. siRNA Transfection
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Paul, E.O.; Kenneth, P.O. Pancreatic cancer: Why is it so hard to treat? Ther. Adv. Gastroenterol. 2013, 6, 321–337. [Google Scholar]
- Sheikh, R.; Walsh, N.; Clynes, M.; O’Connor, R.; McDermott, R. Challenges of drug resistance in the management of pancreatic cancer. Exp. Rev. Anticancer Ther. 2010, 10, 1647–1661. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Yuging, Z.; Xianjun, Y.; Drake, L.; Changyi, C.; Qizhi, Y.; Min, L. Overcoming Drug Resistance in Pancreatic Cancer. Exp. Opin. Ther. Targets. 2011, 15, 817–828. [Google Scholar]
- Comella, P.; Casaretti, R.; Sandomenico, C.; Avallone, A.; Franco, L. Role of oxaliplatin in the treatment of colorectal cancer. Ther. Clin. Risk Manag. 2009, 5, 229–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inadomi, K.; Kusaba, H.; Matsushita, Y.; Tanaka, R.; Mitsugi, K.; Arimizu, K.; Hirano, G.; Makiyama, A.; Ohmura, H.; Uchino, K.; et al. Efficacy and Safety Analysis of Oxaliplatin-based Chemotherapy for Advanced Gastric Cancer. Anticancer Res. 2017, 37, 2663–2671. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef]
- Saung, M.T.; Zheng, L. Current Standards of Chemotherapy for Pancreatic Cancer. Clin. Ther. 2017, 39, 2125–2134. [Google Scholar] [CrossRef] [Green Version]
- Alcindor, T.; Beauger, N. Oxaliplatin: A review in the era of molecularly targeted therapy. Curr. Oncol. 2011, 18, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Seetharam, R.; Sood, A.; Goel, S. Oxaliplatin: Pre-clinical perspectives on the mechanisms of action, response and resistance. Ecancermedicalscience 2009, 3, 153. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Balibrea, E.; Martinez-Cardus, A.; Gines, A.; Ruiz de Porras, V.; Moutinho, C.; Layos, L.; Manzano, J.L.; Buges, C.; Bystrup, S.; Esteller, M.; et al. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol. Cancer Ther. 2015, 14, 1767–1776. [Google Scholar] [CrossRef] [Green Version]
- Rakitina, T.V.; Vasilevskaya, I.A.; O’Dwyer, P.J. Additive interaction of oxaliplatin and 17-allylamino-17-demethoxygeldanamycin in colon cancer cell lines results from inhibition of nuclear factor kappaB signaling. Cancer Res. 2003, 63, 8600–8605. [Google Scholar] [PubMed]
- Martin, L.P.; Hamilton, T.C.; Schilder, R.J. Platinum resistance: The role of DNA repair pathways. Clin. Cancer Res. 2008, 14, 1291–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.H.; Li, C.; Xiao, Z.Q. Proteomics for identifying mechanisms and biomarkers of drug resistance in cancer. J. Proteom. 2011, 74, 2642–2649. [Google Scholar] [CrossRef]
- An, Y.; Zhou, L.; Huang, Z.; Nice, E.C.; Zhang, H.; Huang, C. Molecular insights into cancer drug resistance from a proteomics perspective. Exp. Rev. Proteom. 2019, 16, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Balibrea, E.; Plasencia, C.; Gines, A.; Martinez-Cardus, A.; Musulen, E.; Aguilera, R.; Manzano, J.L.; Neamati, N.; Abad, A. A proteomic approach links decreased pyruvate kinase M2 expression to oxaliplatin resistance in patients with colorectal cancer and in human cell lines. Mol. Cancer Ther. 2009, 8, 771–778. [Google Scholar] [CrossRef] [Green Version]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Basu, A.; Krishnamurthy, S. Cellular responses to Cisplatin-induced DNA damage. J. Nucleic Acids 2010, 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, C.R.R.; Silva, M.M.; Quinet, A.; Cabral-Neto, J.B.; Menck, C.F.M. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics 2018, 73, e478s. [Google Scholar] [CrossRef]
- Zitvogel, L.; Galluzzi, L.; Kepp, O.; Smyth, M.J.; Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015, 15, 405–414. [Google Scholar] [CrossRef]
- Huo, Y.; Zong, Z.; Wang, Q.; Zhang, Z.; Deng, H. ISG15 silencing increases cisplatin resistance via activating p53-mediated cell DNA repair. Oncotarget 2017, 8, 107452–107461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- William-Faltaos, S.; Rouillard, D.; Lechat, P.; Bastian, G. Cell cycle arrest by oxaliplatin on cancer cells. Fundam. Clin. Pharmacol. 2007, 21, 165–172. [Google Scholar] [CrossRef]
- Arango, D.; Wilson, A.J.; Shi, Q.; Corner, G.A.; Aranes, M.J.; Nicholas, C.; Lesser, M.; Mariadason, J.M.; Augenlicht, L.H. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br. J. Cancer 2004, 91, 1931–1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voland, C.; Bord, A.; Peleraux, A.; Penarier, G.; Carriere, D.; Galiegue, S.; Cvitkovic, E.; Jbilo, O.; Casellas, P. Repression of cell cycle-related proteins by oxaliplatin but not cisplatin in human colon cancer cells. Mol. Cancer Ther. 2006, 5, 2149–2157. [Google Scholar] [CrossRef] [Green Version]
- Sarin, N.; Engel, F.; Kalayda, G.V.; Mannewitz, M.; Cinatl, J., Jr.; Rothweiler, F.; Michaelis, M.; Saafan, H.; Ritter, C.A.; Jaehde, U.; et al. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS ONE 2017, 12, e0181081. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Chen, Y.; Saha, M.N.; Chen, J.; Evans, K.; Qiu, L.; Reece, D.; Chen, G.A.; Chang, H. Targeting phospho-MARCKS overcomes drug-resistance and induces antitumor activity in preclinical models of multiple myeloma. Leukemia 2015, 29, 715–726. [Google Scholar] [CrossRef]
- Glaeser, K.; Urban, M.; Fenech, E.; Voloshanenko, O.; Kranz, D.; Lari, F.; Christianson, J.C.; Boutros, M. ERAD-dependent control of the Wnt secretory factor Evi. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Zhong, Z.; Virshup, D. Wnt Signaling and Drug Resistance in Cancer. Mol. Pharmacol. 2020, 97, 72–89. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Fong, L.W.R.; Yu, E.; Wu, R.; Trott, J.F.; Weiss, R.H. Upregulation of MARCKS in kidney cancer and its potential as a therapeutic target. Oncogene 2017, 36, 3588–3598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.H.; Thai, P.; Yoneda, K.; Adler, K.B.; Yang, P.C.; Wu, R. A peptide that inhibits function of Myristoylated Alanine-Rich C Kinase Substrate (MARCKS) reduces lung cancer metastasis. Oncogene 2014, 33, 3696–3706. [Google Scholar] [CrossRef] [Green Version]
- Fong, L.W.R.; Yang, D.C.; Chen, C.H. Myristoylated alanine-rich C kinase substrate (MARCKS): A multirole signaling protein in cancers. Cancer Metastasis Rev. 2017, 36, 737–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.T.; Anastas, J.N.; Toroni, R.A.; Shinohara, M.M.; Goodson, J.M.; Bosserhoff, A.K.; Chien, A.J.; Moon, R.T. WLS inhibits melanoma cell proliferation through the beta-catenin signalling pathway and induces spontaneous metastasis. EMBO Mol. Med. 2012, 4, 1294–1307. [Google Scholar] [CrossRef]
- Adell, T.; Salo, E.; Boutros, M.; Bartscherer, K. Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 2009, 136, 905–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlad-Fiegen, A.; Langerak, A.; Eberth, S.; Muller, O. The Wnt pathway destabilizes adherens junctions and promotes cell migration via beta-catenin and its target gene cyclin D1. FEBS Open Bio 2012, 2, 26–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manic, S.; Gatti, L.; Carenini, N.; Fumagalli, G.; Zunino, F.; Perego, P. Mechanisms controlling sensitivity to platinum complexes: Role of p53 and DNA mismatch repair. Curr. Cancer Drug Targets 2003, 3, 21–29. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef] [PubMed]
- Pfister, N.T.; Prives, C. Transcriptional Regulation by Wild-Type and Cancer-Related Mutant Forms of p53. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Yu, L.; Song, X.; Bi, J.; Jiang, L.; Wang, Y.; He, M.; Xiao, Q.; Sun, M.; Olopade, O.I.; et al. Intrinsic adriamycin resistance in p53-mutated breast cancer is related to the miR-30c/FANCF/REV1-mediated DNA damage response. Cell Death Dis. 2019, 10, 666. [Google Scholar] [CrossRef]
- Blandino, G.; Di Agostino, S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J. Exp. Clin. Cancer Res. 2018, 37, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ma, H.; Wang, Z.; Zhang, S.; Yang, H.; Fang, Z. EZH2 Palmitoylation Mediated by ZDHHC5 in p53-Mutant Glioma Drives Malignant Development and Progression. Cancer Res. 2017, 77, 4998–5010. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Tahaney, W.M.; Mazumdar, A.; Savage, M.I.; Brown, P.H. Molecularly targeted therapies for p53-mutant cancers. Cell Mol. Life Sci. 2017, 74, 4171–4187. [Google Scholar] [CrossRef]
- Yang, L.; Zhou, Y.; Li, Y.; Zhou, J.; Wu, Y.; Cui, Y.; Yang, G.; Hong, Y. Mutations of p53 and KRAS activate NF-kappaB to promote chemoresistance and tumorigenesis via dysregulation of cell cycle and suppression of apoptosis in lung cancer cells. Cancer Lett. 2015, 357, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Fiorini, C.; Cordani, M.; Padroni, C.; Blandino, G.; Di Agostino, S.; Donadelli, M. Mutant p53 stimulates chemoresistance of pancreatic adenocarcinoma cells to gemcitabine. Biochim. Biophys. Acta 2015, 1853, 89–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deer, E.L.; Gonzalez-Hernandez, J.; Coursen, J.D.; Shea, J.E.; Ngatia, J.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 2010, 39, 425–435. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.K.; Jang, M.; Song, M.J.; Kim, D.; Kim, Y.; Jang, H.H. Redox-Mediated Mechanism of Chemoresistance in Cancer Cells. Antioxidants 2019, 8, 471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burhans, W.C.; Heintz, N.H. The cell cycle is a redox cycle: Linking phase-specific targets to cell fate. Free Radic. Biol. Med. 2009, 47, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, F.; Corcos, L.; Durand, S.; Simon, B.; Le Jossic-Corcos, C. Acquisition of anticancer drug resistance is partially associated with cancer stemness in human colon cancer cells. Int. J. Oncol 2016, 49, 2558–2568. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.; Schick, J.; Conway, S.; Biade, S.; Laub, P.B.; Stevenson, J.P.; Hamilton, T.C.; O’Dwyer, P.J.; Johnson, S.W. Identification of genes associated with platinum drug sensitivity and resistance in human ovarian cancer cells. Br. J. Cancer 2005, 92, 1149–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budhwani, M.; Mazzieri, R.; Dolcetti, R. Plasticity of Type I Interferon-Mediated Responses in Cancer Therapy: From Anti-tumor Immunity to Resistance. Front. Oncol. 2018, 8, 322. [Google Scholar] [CrossRef] [Green Version]
- Sistigu, A.; Yamazaki, T.; Vacchelli, E.; Chaba, K.; Enot, D.P.; Adam, J.; Vitale, I.; Goubar, A.; Baracco, E.E.; Remedios, C.; et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 2014, 20, 1301–1309. [Google Scholar] [CrossRef]
- Chen, C.H.; Statt, S.; Chiu, C.L.; Thai, P.; Arif, M.; Adler, K.B.; Wu, R. Targeting myristoylated alanine-rich C kinase substrate phosphorylation site domain in lung cancer. Mechanisms and therapeutic implications. Am. J. Respir. Crit. Care Med. 2014, 190, 1127–1138. [Google Scholar] [CrossRef] [Green Version]
- Hanada, S.; Kakehashi, A.; Nishiyama, N.; Wei, M.; Yamano, S.; Chung, K.; Komatsu, H.; Inoue, H.; Suehiro, S.; Wanibuchi, H. Myristoylated alanine-rich C-kinase substrate as a prognostic biomarker in human primary lung squamous cell carcinoma. Cancer Biomark. 2013, 13, 289–298. [Google Scholar] [CrossRef]
- Ziemba, B.; Burke, J.; Masson, G.; Williams, R.; Falke, J. Regulation of PI3K by PKC and MARCKS: Single-Molecule Analysis of a Reconstituted Signaling Pathway. Biophys. J. 2016, 110, 1811–1825. [Google Scholar] [CrossRef] [Green Version]
- Xiu, P.; Dong, X.; Dong, X.; Xu, Z.; Zhu, H.; Liu, F.; Wei, Z.; Zhai, B.; Kanwar, J.R.; Jiang, H.; et al. Secretory clusterin contributes to oxaliplatin resistance by activating Akt pathway in hepatocellular carcinoma. Cancer Sci. 2013, 104, 375–382. [Google Scholar] [CrossRef]
- Chen, J.; Huang, X.F.; Qiao, L.; Katsifis, A. Insulin caused drug resistance to oxaliplatin in colon cancer cell line HT29. J. Gastrointest. Oncol. 2011, 2, 27–33. [Google Scholar] [CrossRef]
- Leelawat, K.; Narong, S.; Udomchaiprasertkul, W.; Leelawat, S.; Tungpradubkul, S. Inhibition of PI3K increases oxaliplatin sensitivity in cholangiocarcinoma cells. Cancer Cell Int. 2009, 9, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Yu, S.; Sakamori, R.; Stypulkowski, E.; Gao, N. Wntless in Wnt secretion: Molecular, cellular and genetic aspects. Front. Biol. 2012, 7, 587–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Polakis, P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 2007, 17, 45–51. [Google Scholar] [CrossRef]
- Barker, N.; Clevers, H. Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 2006, 5, 997–1014. [Google Scholar] [CrossRef] [PubMed]
- Kukcinaviciute, E.; Jonusiene, V.; Sasnauskiene, A.; Dabkeviciene, D.; Eidenaite, E.; Laurinavicius, A. Significance of Notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT116. J. Cell Biochem. 2018, 119, 5913–5920. [Google Scholar] [CrossRef]
- Chikazawa, N.; Tanaka, H.; Tasaka, T.; Nakamura, M.; Tanaka, M.; Onishi, H.; Katano, M. Inhibition of Wnt signaling pathway decreases chemotherapy-resistant side-population colon cancer cells. Anticancer Res. 2010, 30, 2041–2048. [Google Scholar]
- Cui, J.; Jiang, W.; Wang, S.; Wang, L.; Xie, K. Role of Wnt/beta-catenin signaling in drug resistance of pancreatic cancer. Curr. Pharm. Des. 2012, 18, 2464–2471. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, P.; Yadav, V.; Toftdahl, M.; Andersson, T. WNT5A-Induced Activation of the Protein Kinase C Substrate MARCKS Is Required for Melanoma Cell Invasion. Cancers 2020, 12, 346. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.; Nam, H.; Kim, Y.S.; Moon, M.H. Dual-purpose sample trap for on-line strong cation-exchange chromatography/reversed-phase liquid chromatography/tandem mass spectrometry for shotgun proteomics. Application to the human Jurkat T-cell proteome. J. Chromatogr. A 2005, 1070, 193–200. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.E.; Kim, E.-K.; Song, M.-J.; Kim, T.-Y.; Jang, H.H.; Kang, D. SILAC-Based Quantitative Proteomic Analysis of Oxaliplatin-Resistant Pancreatic Cancer Cells. Cancers 2021, 13, 724. https://doi.org/10.3390/cancers13040724
Kim YE, Kim E-K, Song M-J, Kim T-Y, Jang HH, Kang D. SILAC-Based Quantitative Proteomic Analysis of Oxaliplatin-Resistant Pancreatic Cancer Cells. Cancers. 2021; 13(4):724. https://doi.org/10.3390/cancers13040724
Chicago/Turabian StyleKim, Young Eun, Eun-Kyung Kim, Min-Jeong Song, Tae-Young Kim, Ho Hee Jang, and Dukjin Kang. 2021. "SILAC-Based Quantitative Proteomic Analysis of Oxaliplatin-Resistant Pancreatic Cancer Cells" Cancers 13, no. 4: 724. https://doi.org/10.3390/cancers13040724
APA StyleKim, Y. E., Kim, E. -K., Song, M. -J., Kim, T. -Y., Jang, H. H., & Kang, D. (2021). SILAC-Based Quantitative Proteomic Analysis of Oxaliplatin-Resistant Pancreatic Cancer Cells. Cancers, 13(4), 724. https://doi.org/10.3390/cancers13040724