The Synergism between DHODH Inhibitors and Dipyridamole Leads to Metabolic Lethality in Acute Myeloid Leukemia
Abstract
:Simple Summary
Abstract
1. Introduction
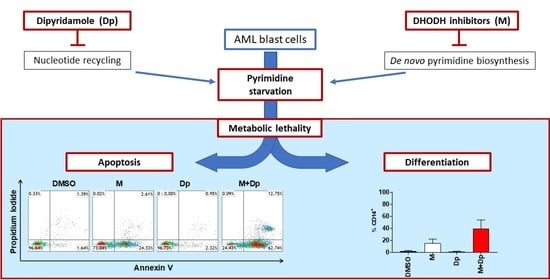
2. Results
2.1. MEDS433 Induces Apoptosis in Several AML Cell Lines
2.2. Apoptosis Is Both the Result of Differentiation and a Direct Effect of DHODH Inhibition
2.3. The Combination of MEDS433 with Classical Antileukemic Agents Results in Near-Additive Effects
2.4. The Combination of MEDS433 with hENT1/2 Inhibitors Results in Synergistic Effects Against AML
2.5. Uridine Reversal: A Matter of Concentration and Transporters
2.6. Approaching the Clinical Setting (I): Effectiveness
2.7. Approaching the Clinical Setting (II): Toxicity
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. Co-Culture
4.4. Primary Cells
4.5. Annexin V and Differentiation Assays
4.6. Flow Cytometry
4.7. Caspase Activity Assay
4.8. Cell Lysis and Western Blot Assay
4.9. RNA Extraction and Gene Expression Analysis
4.10. Cell Staining
4.11. Trypan Blue Staining Assay
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Data and Sample Availability
References
- Sykes, D.B.; Kfoury, Y.S.; Mercier, F.E.; Wawer, M.J.; Law, J.M.; Haynes, M.K.; Lewis, T.A.; Schajnovitz, A.; Jain, E.; Lee, D.; et al. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell 2016, 167, 171–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löffler, M.; Jöckel, J.; Schuster, G. Dihydroorotatubiquinone oxidoreductase links mitochondria in the biosynthesis of pyrimidine nucleotides. Mol. Cell. Biochem. 1997, 174, 125–129. [Google Scholar] [CrossRef]
- Bajzikova, M.; Kovarova, J.; Coelho, A.R.; Boukalova, S.; Oh, S.; Rohlenova, K.; Svec, D.; Hubackova, S.; Endaya, B.; Judasova, K.; et al. Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 2019, 29, 399–416. [Google Scholar] [CrossRef] [Green Version]
- Sainas, S.; Pippione, A.C.; Lupino, E.; Giorgis, M.; Circosta, P.; Gaidano, V.; Goyal, P.; Bonanni, D.; Rolando, B.; Cignetti, A.; et al. Targeting myeloid differentiation using potent 2-hydroxypyrazolo [1, 5-a] pyridine scaffold-based human dihydroorotate dehydrogenase inhibitors. J. Med. Chem. 2018, 61, 6034–6055. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Weetall, M.; Trotta, C.; Cintron, K.; Ma, J.; Kim, M.J.; Furia, B.; Romfo, C.; Graci, J.D.; Li, W.; et al. Targeting of Hematologic Malignancies with PTC299, A Novel Potent Inhibitor of Dihydroorotate Dehydrogenase with Favorable Pharmaceutical Properties. Mol. Cancer Ther. 2019, 18, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Quah, J.Y.; Ng, Y.; Chooi, J.-Y.; Toh, S.H.-M.; Lin, B.; Tan, T.Z.; Hosoi, H.; Osato, M.; Seet, Q.; et al. ASLAN003, a potent dihydroorotate dehydrogenase inhibitor for differentiation of acute myeloid leukemia. Haematologica 2019, 105, 2286–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christian, S.; Merz, C.; Evans, L.; Gradl, S.; Seidel, H.; Friberg, A.; Eheim, A.; Lejeune, P.; Brzezinka, K.; Zim-mermann, K.; et al. The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia 2019, 33, 2403–2415. [Google Scholar] [CrossRef] [Green Version]
- Sykes, D.B. The Emergence of Dihydroorotate Dehydrogenase (DHODH) as a Therapeutic Target in Acute Myeloid Leukemia. Expert. Opin. Ther. Targets 2018, 22, 893–898. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Wang, W.; Chen, W.; Lian, F.; Lang, L.; Huang, Y.; Xu, Y.; Zhang, N.; Chen, Y.; Liu, M.; et al. Pharmacological inhibition of dihydroorotate dehydrogenase induces apoptosis and differentiation in acute myeloid leukemia cells. Haematologica 2018, 103, 1472–1483. [Google Scholar] [CrossRef] [PubMed]
- Sainas, S.; Pippione, A.; Boschi, D.; Gaidano, V.; Circosta, P.; Cignetti, A.; Dosio, F.; Lolli, M. DHODH inhibitors and leukemia: An emergent interest for new myeloid differentiation agents. Drugs Futur. 2018, 43, 823. [Google Scholar] [CrossRef]
- Saunthararajah, Y. Mysteries of partial dihydroorotate dehydrogenase inhibition and leukemia terminal differentiation. Haematologica 2020, 105, 2191–2193. [Google Scholar] [CrossRef] [PubMed]
- Ladds, M.J.G.W.; Van Leeuwen, I.M.M.; Drummond, C.J.; Chu, S.; Healy, A.R.; Popova, G.; Fernandez, A.P.; Mollick, T.; Darekar, S.; Sedimbi, S.K.; et al. A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage. Nat. Commun. 2018, 9, 1107. [Google Scholar] [CrossRef] [Green Version]
- Werner, B.; Gallagher, R.E.; Paietta, E.M.; Litzow, M.R.; Tallman, M.S.; Wiernik, P.H.; Slack, J.L.; Willman, C.L.; Sun, Z.; Traulsen, A.; et al. Dynamics of Leukemia Stem-like Cell Extinction in Acute Promyelocytic Leukemia. Cancer Res. 2014, 74, 5386–5396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodion, P.; Wagener, T.; Stoter, G.; Drozd, A.; Lev, L.; Skovsgaard, T.; Renard, J.; Cavalli, F. Phase II trial with Brequinar (DUP-785, NSC 368390) in patients with metastatic colorectal cancer: A study of the Early Clinical Trials Group of the EORTC. Ann. Oncol. 1990, 1, 79–80. [Google Scholar] [CrossRef]
- Urba, S.G.; Doroshow, J.; Cripps, C.; Robert, F.; Velez-Garcia, E.; Dallaire, B.; Adams, D.; Carlson, R.; Grillo-Lopez, A.; Gyves, J. Multicenter phase II trial of brequinar sodium in patients with advanced squamous-cell carcinoma of the head and neck. Cancer Chemother. Pharmacol. 1992, 31, 167–169. [Google Scholar] [CrossRef]
- Moore, M.; Maroun, J.; Robert, F.; Natale, R.B.; Neidhart, J.; Dallaire, B.; Sisk, R.; Gyves, J. Multicenter phase II study of brequinar sodium in patients with advanced gastrointestinal cancer. Investig. New Drugs 1993, 11, 61–65. [Google Scholar] [CrossRef]
- Okesli, A.; Khosla, C.; Bassik, M.C. Human pyrimidine nucleotide biosynthesis as a target for antiviral chemotherapy. Curr. Opin. Biotechnol. 2017, 48, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, D. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the an-thracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999, 57, 727–741. [Google Scholar] [CrossRef]
- Eom, Y.-W.; Kim, M.A.; Park, S.S.; Goo, M.J.; Kwon, H.J.; Sohn, S.; Kim, W.-H.; Yoon, G.; Choi, K.S. Two distinct modes of cell death induced by doxorubicin: Apoptosis and cell death through mitotic catastrophe accompanied by senescence-like phenotype. Oncogene 2005, 24, 4765–4777. [Google Scholar] [CrossRef] [Green Version]
- Gamen, S.; Anel, A.; Pérez-Galán, P.; Lasierra, P.; Johnson, D.; Piñeiro, A.; Naval, J. Doxorubicin Treatment Activates a Z-VAD-Sensitive Caspase, Which Causes ΔΨm Loss, Caspase-9 Activity, and Apoptosis in Jurkat Cells. Exp. Cell Res. 2000, 258, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Uchiumi, T.; Yagi, M.; Matsumoto, S.; Amamoto, R.; Takazaki, S.; Yamaza, H.; Nonaka, K.; Kang, D. Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci. Rep. 2013, 33, e00021. [Google Scholar] [CrossRef]
- Buolamwini, J.K. ChemInform Abstract: Nucleoside Transport Inhibitors: Structure-Activity Relationships and Potential Therapeutic Applications. Curr. Med. Chem. 2010, 28, 35–66. [Google Scholar] [CrossRef]
- Fairus, A.M.; Choudhary, B.; Hosahalli, S.; Kavitha, N.; Shatrah, O. Dihydroorotate dehydrogenase (DHODH) inhibitors affect ATP depletion, endogenous ROS and mediate S-phase arrest in breast cancer cells. Biochimie 2017, 135, 154–163. [Google Scholar] [CrossRef]
- Dorasamy, M.S.; Choudhary, B.; Nellore, K.; Subramanya, H.; Wong, P.-F. Dihydroorotate dehydrogenase Inhibitors Target c-Myc and Arrest Melanoma, Myeloma and Lymphoma cells at S-phase. J. Cancer 2017, 8, 3086–3098. [Google Scholar] [CrossRef] [Green Version]
- Pizzorno, G.; Cao, D.; Leffert, J.J.; Russell, R.L.; Zhang, D.; Handschumacher, R.E. Homeostatic control of uridine and the role of uridine phosphorylase: A biological and clinical update. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2002, 1587, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Moriwaki, Y.; Takahashi, S.; Tsutsumi, Z.; Ka, T.; Fukuchi, M.; Hada, T. Effect of beer on the plasma concentrations of uridine and purine bases. Metabolism 2002, 51, 1317–1323. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Rausch, C.R.; Benton, C.; Kadia, T.; Jain, N.; Pemmaraju, N.; Daver, N.; Covert, W.; Marx, K.R.; Mace, M.; et al. Clinical experience with the BCL 2-inhibitor venetoclax in combination therapy for relapsed and refractory acute myeloid leukemia and related myeloid malignancies. Am. J. Hematol. 2018, 93, 401–407. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.E.; Redner, R.L. An ATRActive future for differentiation therapy in AML. Blood Rev. 2015, 29, 263–268. [Google Scholar] [CrossRef] [Green Version]
- Stein, E.; Yen, K. Targeted differentiation therapy with mutant IDH inhibitors: Early experiences and parallels with other differentiation agents. Annu. Rev. Cancer Biol. 2017, 1, 379–401. [Google Scholar] [CrossRef]
- Zecchini, V.; Frezza, C. Metabolic synthetic lethality in cancer therapy. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1858, 723–731. [Google Scholar] [CrossRef]
- Gaude, E.; Frezza, C. Defects in mitochondrial metabolism and cancer. Cancer Metab. 2014, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Farber, S.; Diamond, L.K.; Mercer, R.D.; Sylvester, R.F., Jr.; Wolff, J.A. Temporary remissions in acute leu-kemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid (aminopterin). N. Engl. J. Med. 1948, 238, 787–793. [Google Scholar] [CrossRef]
- Brown, K.K.; Spinelli, J.B.; Asara, J.M.; Toker, A. Adaptive reprogramming of de novo pyrimidine syn-thesis is a metabolic vulnerability in triple-negative breast cancer. Cancer Discov. 2017, 7, 391–399. [Google Scholar] [CrossRef] [Green Version]
- Shukla, S.K.; Purohit, V.; Mehla, K.; Gunda, V.; Chaika, N.V.; Vernucci, E.; King, R.J.; Abrego, J.; Goode, G.D.; Dasgupta, A.; et al. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gem-citabine resistance to pancreatic cancer. Cancer Cell 2017, 32, 71–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, T.; Haapa-Paananen, S.; Kaminskyy, V.O.; Kohonen, P.; Fey, V.; Zhivotovsky, B.; Kallioniemi, O.; Perälä, M. Inhibition of the mitochondrial pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase by doxorubicin and brequinar sensitizes cancer cells to TRAIL-induced apoptosis. Oncogene 2014, 33, 3538–3549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imanishi, S.; Takahashi, R.; Katagiri, S.; Kobayashi, C.; Umezu, T.; Ohyashiki, K.; Ohyashiki, J.H. Teriflunomide restores 5-azacytidine sensitivity via activation of pyrimidine salvage in 5-azacytidine-resistant leukemia cells. Oncotarget 2017, 8, 69906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dexter, D.L.; Hesson, D.P.; Ardecky, R.J.; Rao, G.V.; Tippett, D.L.; Dusak, B.A.; Paull, K.D.; Plowman, J.; DeLarco, B.M.; Narayanan, V.L.; et al. Activity of a novel 4-quinolinecarboxylic acid, NSC 368390 [6-fluoro-2-(2′-fluoro-1, 1′-biphenyl-4-yl)-3-methyl-4-quinolinecarboxylic acid sodium salt], against experimental tumors. Cancer Res. 1985, 45, 5563–5568. [Google Scholar]
- Madak, J.T.; Bankhead, A., III; Cuthbertson, C.R.; Showalter, H.D.; Neamati, N. Revisiting the role of di-hydroorotate dehydrogenase as a therapeutic target for cancer. Pharmacol. Ther. 2019, 195, 111–131. [Google Scholar] [CrossRef]
- Karle, J.M.; Anderson, L.W.; Dietrick, D.D.; Cysyk, R.L. Determination of serum and plasma uridine levels in mice, rats, and humans by high-pressure liquid chromatography. Anal. Biochem. 1980, 109, 41–46. [Google Scholar] [CrossRef]
- Xiong, R.; Zhang, L.; Li, S.; Sun, Y.; Ding, M.; Wang, Y.; Zhao, Y.; Wu, Y.; Shang, W.; Jiang, X.; et al. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2. Protein Cell 2020, 11, 723–739. [Google Scholar] [CrossRef]
- Li, Z.; Philip, M.; Ferrell, P.B. Alterations of T-cell-mediated immunity in acute myeloid leukemia. Oncogene 2020, 39, 3611–3619. [Google Scholar] [CrossRef]
- Peters, G.J.; Kraal, I.; Pinedo, H.M. In vitro and in vivo studies on the combination of Brequinar sodium (DUP-785; NSC 368390) with 5-fluorouracil; effects of uridine. Br. J. Cancer 1992, 65, 229–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markman, M.; Chan, T.C.K.; Cleary, S.; Howell, S.B. Phase I trial of combination therapy of cancer with N-phosphanacetyl-L-aspartic acid and dipyridamole. Cancer Chemother. Pharmacol. 1987, 19, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Carrà, G.; Lingua, M.F.; Maffeo, B.; Guerrasio, A.; Taulli, R.; Morotti, A. PF372 inhibition of bromodomain and extra-terminal (bet) proteins increases sensitivity to venetoclax in chronic lymphocytic leukemia. HemaSphere 2019, 3, 137. [Google Scholar] [CrossRef]
- Carrà, G.; Panuzzo, C.; Torti, D.; Parvis, G.; Crivellaro, S.; Familiari, U.; Volante, M.; Morena, D.; Lingua, M.F.; Brancaccio, M.; et al. Therapeutic inhibition of USP7-PTEN network in chronic lymphocytic leukemia: A strategy to overcome TP53 mutated/deleted clones. Oncotarget 2017, 8, 35508–35522. [Google Scholar] [CrossRef] [PubMed] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaidano, V.; Houshmand, M.; Vitale, N.; Carrà, G.; Morotti, A.; Tenace, V.; Rapelli, S.; Sainas, S.; Pippione, A.C.; Giorgis, M.; et al. The Synergism between DHODH Inhibitors and Dipyridamole Leads to Metabolic Lethality in Acute Myeloid Leukemia. Cancers 2021, 13, 1003. https://doi.org/10.3390/cancers13051003
Gaidano V, Houshmand M, Vitale N, Carrà G, Morotti A, Tenace V, Rapelli S, Sainas S, Pippione AC, Giorgis M, et al. The Synergism between DHODH Inhibitors and Dipyridamole Leads to Metabolic Lethality in Acute Myeloid Leukemia. Cancers. 2021; 13(5):1003. https://doi.org/10.3390/cancers13051003
Chicago/Turabian StyleGaidano, Valentina, Mohammad Houshmand, Nicoletta Vitale, Giovanna Carrà, Alessandro Morotti, Valerio Tenace, Stefania Rapelli, Stefano Sainas, Agnese Chiara Pippione, Marta Giorgis, and et al. 2021. "The Synergism between DHODH Inhibitors and Dipyridamole Leads to Metabolic Lethality in Acute Myeloid Leukemia" Cancers 13, no. 5: 1003. https://doi.org/10.3390/cancers13051003
APA StyleGaidano, V., Houshmand, M., Vitale, N., Carrà, G., Morotti, A., Tenace, V., Rapelli, S., Sainas, S., Pippione, A. C., Giorgis, M., Boschi, D., Lolli, M. L., Cilloni, D., Cignetti, A., Saglio, G., & Circosta, P. (2021). The Synergism between DHODH Inhibitors and Dipyridamole Leads to Metabolic Lethality in Acute Myeloid Leukemia. Cancers, 13(5), 1003. https://doi.org/10.3390/cancers13051003