CNV Hotspots in Testicular Seminoma Tissue and Seminal Plasma
Abstract
:Simple Summary
Abstract
1. Introduction
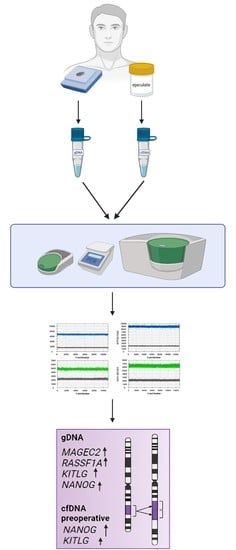
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection
2.2.1. Ejaculate
2.2.2. Seminoma Tissue
2.3. DNA Isolation and Quantification
2.4. Digital Droplet PCR (ddPCR)
2.5. Statistical Analysis
3. Results
3.1. CNV in Tissue Samples
3.2. CNV in cfDNA from Seminal Plasma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lobo, J.; Gillis, A.J.M.; Jerónimo, C.; Henrique, R.; Looijenga, L.H.J. Human germ cell tumors are developmental cancers: Impact of epigenetics on pathobiology and clinic. Int. J. Mol. Sci. 2019, 20, 258. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Moul, J.W.; Schanne, F.J.; Thompson, I.M.; Frazier, H.A.; Peretsman, S.A.; Wettlaufer, J.N.; Rozanski, T.A.; Stack, R.S.; Kreder, K.J.; Hoffman, K.J. Testicular cancer in blacks. A multicenter experience. Cancer 1994, 73, 388–393. [Google Scholar] [CrossRef]
- Baroni, T.; Arato, I.; Mancuso, F.; Calafiore, R.; Luca, G. On the Origin of Testicular Germ Cell Tumors: From Gonocytes to Testicular Cancer. Front. Endocrinol. 2019, 10, 343. [Google Scholar] [CrossRef] [PubMed]
- Raos, D.; Krasic, J.; Masic, S.; Abramovic, I.; Coric, M.; Kruslin, B.; Katusic Bojanac, A.; Bulic-Jakus, F.; Jezek, D.; Ulamec, M.; et al. In Search of TGCT Biomarkers: A Comprehensive in Silico and Histopathological Analysis. Dis. Markers 2020, 2020, 8841880. [Google Scholar] [CrossRef]
- Skakkebæk, N.E.; Rajpert-De Meyts, E.; Main, K.M. Testicular dysgenesis syndrome: An increasingly common developmental disorder with environmental aspects. Hum. Reprod. 2001, 16, 972–978. [Google Scholar] [CrossRef]
- Dong, C.; Hemminki, K. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int. J. Cancer 2001, 92, 144–150. [Google Scholar] [CrossRef]
- Gundy, S.; Babosa, M.; Baki, M.; Bodrogi, I. Increased predisposition to cancer in brothers and offspring of testicular tumor patients. Pathol. Oncol. Res. 2004, 10, 197–203. [Google Scholar] [CrossRef] [Green Version]
- Looijenga, L.H.J.; Zafarana, G.; Grygalewicz, B.; Summersgill, B.; Debiec-Rychter, M.; Veltman, J.; Schoenmakers, E.F.P.M.; Rodriguez, S.; Jafer, O.; Clark, J.; et al. Role of gain of 12p in germ cell tumour development. Apmis 2003, 111, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Kanetsky, P.A.; Mitra, N.; Vardhanabhuti, S.; Li, M.; David, J.; Letrero, R.; Ciosek, S.L.; Doody, D.R.; Lauren, M.; Weaver, J.; et al. Common variation in KITLG and at 5q31.3 proximate to SPRY4 predispose to testicular germ cell cancer Peter. Nat. Genet. 2010, 41, 811–815. [Google Scholar] [CrossRef] [Green Version]
- Turnbull, C.; Rapley, E.A.; Seal, S.; Pernet, D.; Renwick, A.; Hughes, D.; Ricketts, M.; Linger, R.; Nsengimana, J.; Deloukas, P.; et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat. Genet. 2010, 42, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Rapley, E.A.; Turnbull, C.; Al Olama, A.A.; Dermitzakis, E.T.; Linger, R.; Huddart, R.A.; Renwick, A.; Hughes, D.; Hines, S.; Seal, S.; et al. A genome-wide association study of testicular germ cell tumor. Nat. Genet. 2009, 41, 807–810. [Google Scholar] [CrossRef] [Green Version]
- Macé, A.; Kutalik, Z.; Valsesia, A. Copy number variation. Methods Mol. Biol. 2018, 1793, 231–258. [Google Scholar] [PubMed]
- Furuya, T.; Suehiro, Y.; Namiki, Y.; Sasaki, K. CNVs Associated with Susceptibility to Cancers: A Mini-Review. J. Cancer Ther. 2015, 06, 413–422. [Google Scholar] [CrossRef] [Green Version]
- Lafin, J.T.; Bagrodia, A.; Woldu, S.; Amatruda, J.F. New insights into germ cell tumor genomics. Andrology 2019, 7, 507–515. [Google Scholar] [CrossRef] [Green Version]
- Haugen, T.B. Functions of genes related to testicular germ cell tumour development. Andrology 2019, 7, 527–535. [Google Scholar]
- Raos, D.; Ulamec, M.; Katusic Bojanac, A.; Bulic-Jakus, F.; Jezek, D.; Sincic, N. Epigenetically inactivated RASSF1A as a tumor biomarker. Bosn. J. Basic Med. Sci. 2020, 21, 386–397. [Google Scholar] [CrossRef]
- LeBron, C.; Pal, P.; Brait, M.; Dasgupta, S.; Guerrero-Preston, R.; Looijenga, L.H.J.; Kowalski, J.; Netto, G.; Hoque, M.O. Genome-wide analysis of genetic alterations in testicular primary seminoma using high resolution single nucleotide polymorphism arrays. Genomics 2011, 97, 341–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dieckmann, K.P.; Simonsen-Richter, H.; Kulejewski, M.; Anheuser, P.; Zecha, H.; Isbarn, H.; Pichlmeier, U. Serum Tumour Markers in Testicular Germ Cell Tumours: Frequencies of Elevated Levels and Extents of Marker Elevation Are Significantly Associated with Clinical Parameters and with Response to Treatment. Biomed. Res. Int. 2019, 2019, 5030349. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef]
- Ponti, G.; Maccaferri, M.; Micali, S.; Manfredini, M.; Milandri, R.; Bianchi, G.; Pellacani, G.; Kaleci, S.; Chester, J.; Conti, A.; et al. Seminal cell free DNA concentration levels discriminate between prostate cancer and benign prostatic hyperplasia. Anticancer Res. 2018, 38, 5121–5125. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, F.R.; Abramović, I.; Cuenin, C.; Carreira, C. A protocol for good quality genomic DNA isolation from formalin-fixed paraffin-embedded tissues without using commercial kits. bioRxiv 2021. [Google Scholar] [CrossRef]
- Krasic, J.; Abramovic, I.; Vrtaric, A.; Gabaj, N.N. Impact of Preanalytical and Analytical Methods on Cell-Free DNA Diagnostics. Front. Cell Dev. Biol. 2021, 9, 686149. [Google Scholar]
- Laguna, M.P.; Albers, P.; Algaba, F.; Bokemeyer, C.; Boormans, J.L.; Fischer, S.; Fizazi, K.; Gremmels, H.; Leão, R.; Nicolai, N.; et al. EAU Guidelines on Testicular Cancer 2020. Eur. Assoc. Urol. Guidel. 2020. Available online: https://uroweb.org/guideline/testicular-cancer/ (accessed on 28 December 2021).
- Krepischi, A.C.V.; Pearson, P.L.; Rosenberg, C. Germline copy number variations and cancer predisposition. Futur. Oncol. 2012, 8, 441–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraggerud, S.M.; Skotheim, R.I.; Szymanska, J.; Eknæs, M.; Fosså, S.D.; Stenwig, A.E.; Peltomäki, P.; Lothe, R.A. Genome profiles of familial/bilateral and sporadic testicular germ cell tumors. Genes Chromosom. Cancer 2002, 34, 168–174. [Google Scholar] [CrossRef]
- Roelofs, H.; Mostert, M.C.; Pompe, K.; Zafarana, G.; Van Oorschot, M.; Van Gurp, R.J.H.L.M.; Gillis, A.J.M.; Stoop, H.; Beverloo, B.; Oosterhuis, J.W.; et al. Restricted 12p amplification and RAS mutation in human germ cell tumors of the adult testis. Am. J. Pathol. 2000, 157, 1155–1166. [Google Scholar] [CrossRef] [Green Version]
- Becher, R.; Korn, W.M.; Prescher, G. Use of fluorescence in situ hybridization and comparative genomic hybridization in the cytogenetic analysis of testicular germ cell tumors and uveal melanomas. Cancer Genet. Cytogenet. 1997, 93, 22–28. [Google Scholar] [CrossRef]
- Bergthorsson, J.T.; Agnarsson, B.A.; Gudbjartsson, T.; Magnusson, K.; Thoroddsen, A.; Palsson, B.; Bjornsson, J.; Stefansson, K.; Gulcher, J.; Einarsson, G.V.; et al. A genome-wide study of allelic imbalance in human testicular germ cell tumors using microsatellite markers. Cancer Genet. Cytogenet. 2006, 164, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Korn, W.M.; Olde Weghuis, D.E.M.; Suijkerbuijk, R.F.; Schmidt, U.; Otto, T.; Du Manoir, S.; Van Kessel, A.G.; Harstrick, A.; Seeber, S.; Becher, R. Detection of chromosomal DNA gains and losses in testicular germ cell tumors by comparative genomic hybridization. Genes Chromosom. Cancer 1996, 17, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Hoff, A.M.; Kraggerud, S.M.; Alagaratnam, S.; Berg, K.C.G.; Johannessen, B.; Holand, M.; Nilsen, G.; Lingj Rde, O.C.; Andrews, P.W.; Lothe, R.A.; et al. Frequent copy number gains of SLC2A3 and ETV1 in testicular embryonal carcinomas. Endocr. Relat. Cancer 2020, 27, 457–468. [Google Scholar] [CrossRef]
- Rocca, M.S.; Di Nisio, A.; Marchiori, A.; Ghezzi, M.; Opocher, G.; Foresta, C.; Ferlin, A. Copy number variations of E2F1: A new genetic risk factor for testicular cancer. Endocr. Relat. Cancer 2017, 24, 119–125. [Google Scholar] [CrossRef]
- Stadler, Z.K.; Esposito, D.; Shah, S.; Vijai, J.; Yamrom, B.; Levy, D.; Lee, Y.H.; Kendall, J.; Leotta, A.; Ronemus, M.; et al. Rare de novo germline copy-number variation in testicular cancer. Am. J. Hum. Genet. 2012, 91, 379–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018, 11, 3392–3406. [Google Scholar] [CrossRef]
- Cutcutache, I.; Suzuki, Y.; Tan, I.B.; Ramgopal, S.; Zhang, S.; Ramnarayanan, K.; Gan, A.; Lee, H.H.; Tay, S.T.; Ooi, A.; et al. Exome-wide sequencing shows low mutation rates and identifies novel mutated genes in seminomas. Eur. Urol. 2015, 68, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Ponti, G.; Manfredini, M.; Tomasi, A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit. Rev. Oncol. Hematol. 2019, 141, 36–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Struijk, R.B.; Dorssers, L.C.J.; Henneman, P.; Rijlaarsdam, M.A.; Venema, A.; Jongejan, A.; Mannens, M.M.A.M.; Looijenga, L.H.J.; Repping, S.; van Pelt, A.M.M. Comparing genome-scale DNA methylation and CNV marks between adult human cultured ITGA6+ testicular cells and seminomas to assess in vitro genomic stability. PLoS ONE 2020, 15, e0230253. [Google Scholar] [CrossRef]
- Cheng, L.; Albers, P.; Berney, D.M.; Feldman, D.R.; Daugaard, G.; Gilligan, T.; Looijenga, L.H. Testicular Cancer. Nat. Rev. Dis. Prim. 2018, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Skotheim, R.I.; Autio, R.; Lind, G.E.; Kraggerud, S.M.; Andrews, P.W.; Monni, O.; Kallioniemi, O.; Lothe, R.A. Novel genomic aberrations in testicular germ cell tumors by array-CGH, and associated gene expression changes. Cell. Oncol. 2006, 28, 315–326. [Google Scholar] [CrossRef]
- Batool, A.; Karimi, N.; Wu, X.N.; Chen, S.R.; Liu, Y.X. Testicular germ cell tumor: A comprehensive review. Cell. Mol. Life Sci. 2019, 76, 1713–1727. [Google Scholar] [CrossRef]
- Cardoso, H.J.; Figueira, M.I.; Socorro, S. The stem cell factor (SCF)/c-KIT signalling in testis and prostate cancer. J. Cell Commun. Signal. 2017, 11, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Costa, A.L.; Moreira-Barbosa, C.; Lobo, J.; Vilela-Salgueiro, B.; Cantante, M.; Guimarães, R.; Lopes, P.; Braga, I.; Oliveira, J.; Antunes, L.; et al. DNA methylation profiling as a tool for testicular germ cell tumors subtyping. Epigenomics 2018, 10, 1511–1523. [Google Scholar] [CrossRef] [PubMed]
- Bode, P.K.; Barghorn, A.; Fritzsche, F.R.; Riener, M.O.; Kristiansen, G.; Knuth, A.; Moch, H. MAGEC2 is a sensitive and novel marker for seminoma: A tissue microarray analysis of 325 testicular germ cell tumors. Mod. Pathol. 2011, 24, 829–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Pizio, P.; Celton, N.; Menoud, P.A.; Belloc, S.; Cohen Bacrie, M.; Belhadri-Mansouri, N.; Rives, N.; Cabry, R.; Benkhalifa, M. Seminal cell-free DNA and sperm characteristic’s: An added biomarker for male infertility investigation. Andrologia 2021, 53, e13822. [Google Scholar] [CrossRef]
- Neto, F.T.L.; Bach, P.V.; Najari, B.B.; Li, P.S.; Goldstein, M. Spermatogenesis in humans and its affecting factors. Semin. Cell Dev. Biol. 2016, 59, 10–26. [Google Scholar] [CrossRef]
- Edsgärd, D.; Scheel, M.; Hansen, N.T.; Ralfkiaer, U.; Jensen, T.S.; Skakkebæk, N.E.; Brunak, S.; Gupta, R.; Rajpert-De Meyts, E.; Ottesen, A.M. Heterozygous deletion at the RLN1 locus in a family with testicular germ cell cancer identified by integrating copy number variation data with phenome and interactome information. Int. J. Androl. 2011, 34, e122–e123. [Google Scholar] [CrossRef] [PubMed]
- Edsgärd, D.; Dalgaard, M.D.; Weinhold, N.; Wesolowska-Andersen, A.; De Meyts, E.R.; Ottesen, A.M.; Juul, A.; Skakkebæk, N.E.; Jensen, T.S.; Gupta, R.; et al. Genome-wide assessment of the association of rare and common copy number variations to testicular germ cell cancer. Front. Endocrinol. 2013, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Pös, O.; Radvanszky, J.; Buglyó, G.; Pös, Z.; Rusnakova, D.; Nagy, B.; Szemes, T. DNA copy number variation: Main characteristics, evolutionary and pathological aspects. Biomed. J. 2021, 44, 548–559. [Google Scholar] [CrossRef]


| Gene | Location |
|---|---|
| RASSF1A | 3p21.31 |
| NANOG | 12p13.31 |
| KITLG | 12q21.32 |
| MAGEC2 | Xq27.2 |
| Gene | Primer | Sequence of the In-House Assay | Temperature (°C) | No. of Cycles |
| KITLG | F | 5′-GCGGGACTTGGGTCTCATTT-3′ | 57.5 | 40 |
| R | 5′-TCTGGAGCCATGCAAATGGT-3′ | |||
| Gene | Commercial Assay ID | Temperature (°C) | No. of Cycles | |
| RASSF1A | dHsaCNS143255910 | 57.5 | 40 | |
| NANOG | dHsaCNS193219338 | 57.5 | 40 | |
| MAGEC2 | dHsaCNS241647353 | 57.5 | 40 | |
| Clinicopathological Variables | SE Patients (n = 24) | Healthy Volunteers (n = 35) | |
|---|---|---|---|
| Median age, years (range) | 35 (20–49) | 26 (16–42) | |
| TNM classification | T1 | 17 | - |
| T2 | 5 | ||
| T3 | 1 | ||
| Median tumour size (range) cm | 3.4 (0.3–8) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raos, D.; Abramović, I.; Tomić, M.; Vrtarić, A.; Kuliš, T.; Ćorić, M.; Ulamec, M.; Katušić Bojanac, A.; Ježek, D.; Sinčić, N. CNV Hotspots in Testicular Seminoma Tissue and Seminal Plasma. Cancers 2022, 14, 189. https://doi.org/10.3390/cancers14010189
Raos D, Abramović I, Tomić M, Vrtarić A, Kuliš T, Ćorić M, Ulamec M, Katušić Bojanac A, Ježek D, Sinčić N. CNV Hotspots in Testicular Seminoma Tissue and Seminal Plasma. Cancers. 2022; 14(1):189. https://doi.org/10.3390/cancers14010189
Chicago/Turabian StyleRaos, Dora, Irena Abramović, Miroslav Tomić, Alen Vrtarić, Tomislav Kuliš, Marijana Ćorić, Monika Ulamec, Ana Katušić Bojanac, Davor Ježek, and Nino Sinčić. 2022. "CNV Hotspots in Testicular Seminoma Tissue and Seminal Plasma" Cancers 14, no. 1: 189. https://doi.org/10.3390/cancers14010189
APA StyleRaos, D., Abramović, I., Tomić, M., Vrtarić, A., Kuliš, T., Ćorić, M., Ulamec, M., Katušić Bojanac, A., Ježek, D., & Sinčić, N. (2022). CNV Hotspots in Testicular Seminoma Tissue and Seminal Plasma. Cancers, 14(1), 189. https://doi.org/10.3390/cancers14010189