Three-Dimensional Organotypic Cultures Reshape the microRNAs Transcriptional Program in Breast Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
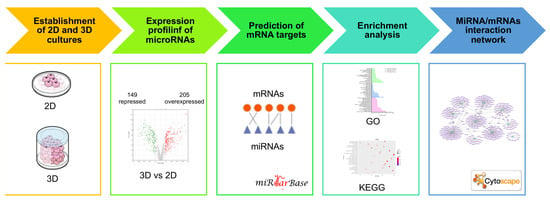
2. Materials and Methods
2.1. Cell Cultures
2.2. Three-Dimensional Organotypic Cultures
2.3. Immunofluorescence Experiments in 3D Cultures
2.4. RNA Isolation
2.5. microRNAs Microarray Analysis
2.6. Target Gene Predictions
2.7. Enrichment Analysis
2.8. Analysis of the miRNA/mRNA Networks
2.9. Statistical Analysis
3. Results
3.1. Establishment of 3D Organotypic Cultures of Breast Cancer Cell Lines
3.2. The Morphology of 3D Organotypic Cultures of Hs578T Breast Cancer Cells
3.3. Differential Expression Profiling of microRNAs in 3D Cultures Compared to 2D Cultures of Hs578T Triple-Negative Breast Cancer Cells
3.4. The Enrichment Analysis of Predicted Target Genes for miRNAs Deregulated in 3D Organotypic Cultures Using GO and KEGG Tools
3.5. Networks of miRNA/mRNA Interactions in 3D Organotypic Cell Cultures
3.6. The Expression Levels of Several miRNAs Modulated in Hs578T 3D Organotypic Cultures Correlate with Expression Found in Tumors of Breast Cancer Patients
3.7. Low Expression of miR-935 and High Levels of Its Target mRNA HIF1A Correlate with Poor Patient Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017, 389, 2430–2442. [Google Scholar] [CrossRef] [Green Version]
- Anders, C.; Carey, L.A. Understanding and treating triple-negative breast cancer. Oncology 2008, 22, 1233–1243. [Google Scholar] [PubMed]
- Yao, H.; He, G.; Yan, S.; Chen, C.; Song, L.; Rosol, T.J.; Deng, X. Triple-negative breast cancer: Is there a treatment on the horizon? Oncotarget 2017, 8, 1913–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, S.; Peyman, S.; Speirs, V. Current and Emerging 3D Models to Study Breast Cancer. Adv. Exp. Med. Biol. 2019, 1152, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Koedoot, E.; Wolters, L.; Smid, M.; Stoilov, P.; Burger, G.A.; Herpers, B.; Yan, K.; Price, L.S.; Martens, J.W.M.; Le Dévédec, S.E.; et al. Differential reprogramming of breast cancer subtypes in 3D cultures and implications for sensitivity to targeted therapy. Sci. Rep. 2021, 11, 7259. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 17, 1033–1055. [Google Scholar] [CrossRef]
- Calin, G.; Croce, C. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Lerner, R.G.; Petritsch, C. A microRNA-operated switch of asymmetric-to-symmetric cancer stem cell divisions. Nat. Cell Biol. 2014, 16, 212–214. [Google Scholar] [CrossRef] [Green Version]
- Pencheva, N.; Tavazoie, S.F. Control of metastatic progression by microRNA regulatory networks. Nat. Cell Biol. 2013, 15, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Bouchie, A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013, 31, 577. [Google Scholar] [CrossRef]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.W.; Simon, R.M. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 2003, 19, 2448–2455. [Google Scholar] [CrossRef] [Green Version]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, N.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Lee, G.Y.; Kenny, P.A.; Lee, E.H.; Bissell, M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 2007, 4, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Härmä, V.; Schukov, H.P.; Happonen, A.; Ahonen, I.; Virtanen, J.; Siitari, H.; Åkerfelt, M.; Lötjönen, J.; Nees, M. Quantification of dynamic morphological drug responses in 3D organotypic cell cultures by automated image analysis. PLoS ONE 2014, 9, e96426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchi, M.; Piperigkou, Z.; Riti, E.; Masola, V.; Onisto, M.; Karamanos, N.K. Long filopodia and tunneling nanotubes define new phenotypes of breast cancer cells in 3D cultures. Matrix Biol. Plus 2020, 6, 100026. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Fang, Q.; Ge, X. Role and Mechanism of mir-5189-3p in Deep Vein Thrombosis of Lower Extremities. Ann. Vasc. Surg. 2021, 77, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zeng, Q.; Qiu, J.; Pang, T.; Ye, F.; Huang, L.; Zhang, X. MiR-183-5p Promotes Proliferation, Metastasis and Angiogenesis in Breast Cancer Cells through Negatively Regulating Four and a Half LIM Protein 1. J. Breast Cancer 2020, 23, 355. [Google Scholar] [CrossRef]
- Cheng, Y.; Xiang, G.; Meng, Y.; Dong, R. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod. Biol. 2016, 16, 225–233. [Google Scholar] [CrossRef]
- Yang, M.; Cui, G.; Ding, M.; Yang, W.; Liu, Y.; Dai, D.; Chen, L. miR-935 promotes gastric cancer cell proliferation by targeting SOX7. Biomed. Pharmacother. 2016, 79, 153–158. [Google Scholar] [CrossRef]
- Huang, G.; Chen, J.; Liu, J.; Zhang, X.; Duan, H.; Fang, Q. MiR-935/HIF1α Feedback Loop Inhibits the Proliferation and Invasiveness of Glioma. OncoTargets Ther. 2020, 13, 10817. [Google Scholar] [CrossRef]
- Chhabra, R. let-7i-5p, miR-181a-2-3p and EGF/PI3K/SOX2 axis coordinate to maintain cancer stem cell population in cervical cancer. Sci. Rep. 2018, 8, 7840. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Geng, D.; Li, S.; Chen, Z.; Sun, M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018, 7, 842–855. [Google Scholar] [CrossRef]
- Bai, X.; Han, G.; Liu, Y.; Jiang, H.; He, Q. MiRNA-20a-5p promotes the growth of triple-negative breast cancer cells through targeting RUNX3. Biomed. Pharmacother. 2018, 103, 1482–1489. [Google Scholar] [CrossRef]
- Wang, Y.; Tao, B.; Li, J.; Mao, X.; He, W.; Chen, Q. Melatonin Inhibits the Progression of Oral Squamous Cell Carcinoma via Inducing miR-25-5p Expression by Directly Targeting NEDD9. Front. Oncol. 2020, 10, 543591. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Wang, D.; Zhang, M.; Deng, Y.; Jiang, H.; Li, Y. miR-615-3p promotes the epithelial-mesenchymal transition and metastasis of breast cancer by targeting PICK1/TGFBRI axis. Exp. Clin. Cancer Res. 2020, 39, 71. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Zhang, M.; Lu, M.M.; Qu, L.Y.; Xu, S.G.; Li, Y.Z.; Wang, M.Y.; Zhu, H.F.; Zhang, Z.Y.; He, G.Y.; et al. EPAS1 targeting by miR-152-3p in Paclitaxel-resistant Breast Cancer. J. Cancer 2020, 11, 5822. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Wang, D.; Kong, Q.; Gao, W.; Sun, J. Function of miR-152 as a Tumor Suppressor in Human Breast Cancer by Targeting PIK3CA. Oncol. Res. 2017, 25, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, B.; Wang, Y.; Chen, G.; Lian, Q.; Wang, H. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol. Lett. 2019, 17, 3835–3841. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Gromisch, C.; Ozturk, S.; Papageorgis, P.; Abdolmaleky, H.M.; Reinhard, B.M.; Thiagalingam, A.; Thiagalingam, S. MicroRNA-4417 is a tumor suppressor and prognostic biomarker for triple-negative breast cancer. Cancer Biol. Ther. 2019, 20, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Toyama, T.; Takahashi, S.; Yoshimoto, N.; Iwasa, M.; Asano, T.; Fujii, Y.; Yamashita, H. miR-1290 and its potential targets are associated with characteristics of estrogen receptor α-positive breast. Endocr. Relat. Cancer 2013, 20, 91–102. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.J.; Liu, Y.H.; Si, J.M.; Ni, R.; Wang, J. Overexpression of miR-1290 contributes to cell proliferation and invasion of non small cell lung cancer by targeting interferon regulatory factor 2. Int. J. Biochem. Cell Biol. 2018, 95, 113–120. [Google Scholar] [CrossRef]
- Yang, Y.; Yin, Z.X.; Wang, Z.Y.; Tian, S.B.; Wang, H.C.; Zhang, F.X.; Li, L.P.; Zheng, C.; Kong, S. miR-7641 depletion suppresses proliferation of gastric cancer cells by targeting ARID1A. Anticancer Drugs 2020, 31, 368–376. [Google Scholar] [CrossRef]
- Yan, Z.; Hong, S.; Song, Y.; Bi, M. microR-4449 Promotes Colorectal Cancer Cell Proliferation via Regulation of SOCS3 and Activation of STAT3 Signaling. Cancer Manag. Res. 2021, 13, 3029–3039. [Google Scholar] [CrossRef]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Qi, A.; Han, J.; Jia, F.; Liu, C. miR-3175 and miR-134 affect proliferation, invasion and apoptosis of glioma cells through PI3K/AKT signaling pathway. J BUON 2019, 24, 2465–2474. [Google Scholar] [PubMed]
- Tang, T.; Yang, Z.; Zhu, Q.; Wu, Y.; Sun, K.; Alahdal, M.; Zhang, Y.; Xing, Y.; Shen, Y.; Xia, T.; et al. Up-regulation of miR-210 induced by a hypoxic microenvironment promotes breast cancer stem cells metastasis, proliferation, and self-renewal by targeting E-cadherin. FASEB J. 2018, 32, 6965–6981. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Nagy, Á.; Bottai, G.; Munkácsy, G.; Szabó, A.; Santarpia, L.; Győrffy, B. miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 2016, 160, 439–446. [Google Scholar] [CrossRef]
- Baker, B.M.; Chen, C. Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J. Cell Sci. 2012, 125, 3015–3024. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.; Li, C.; Lin, Z.; Zhuang, Y.; Flemington, E.K.; Burow, M.E.; Lin, Y.I.; Shan, B. The microRNA expression associated with morphogenesis of breast cancer cells in three-dimensional organotypic culture. Oncol. Rep. 2012, 28, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef]
- Funamoto, K.; Zervantonakis, I.K.; Liu, Y.; Ochs, C.J.; Kim, C.; Kamm, R.D. A novel microfluidic platform for high-resolution imaging of a three-dimensional cell culture under a controlled hypoxic environment. Lab Chip 2012, 12, 4855–4863. [Google Scholar] [CrossRef] [Green Version]
- Colombo, E.; Cattaneo, M.G. Multicellular 3D Models to Study Tumour-Stroma Interactions. Int. J. Mol. Sci. 2021, 22, 1633. [Google Scholar] [CrossRef]
- Peng, B.; Li, C.; Cai, P.; Yu, L.; Zhao, B.; Chen, G. Knockdown of miR-935 increases paclitaxel sensitivity via regulation of SOX7 in non-small-cell lung cancer. Mol. Med. Rep. 2018, 18, 3397–3402. [Google Scholar] [CrossRef]









| Downregulated miRNA | Fold Change | p-Value | Target | Function | Reference |
| miR-5189-3p | −26.31 | 6.61 × 10−7 | JAG1 | Apoptosis | [23] |
| miR-3160-1 | −25.52 | 9.72 × 10−9 | unknown | unknown | N/A |
| miR-183-5p | −21 | 1.03 × 10−7 | FHL1, PDCD4 | Proliferation, metastasis, and angiogenesis | [24,25] |
| miR-935 | −19.58 | 8.67 × 10−8 | SOX7, HIF1A | Proliferation, invasiveness | [26,27] |
| miR-181a-2-3p | −18.72 | 2.62 × 10−7 | EGF, PI3K, SOX2 | Maintain cancer stem cell | [28] |
| miR-20a-5p | −18.66 | 1.80 × 10−9 | HMGA2, RUNX3 | Cell growth, cell mobility, and apoptosis | [29,30] |
| miR-25-5p | −16.26 | 6.75 × 10−8 | NEDD9 | Proliferation, invasion, and migration | [31] |
| miR-615-3p | −15.57 | 4.96 × 10−7 | PICK1/TGFBRI | Promotes the EMT | [32] |
| miR-152-3p | −14.88 | 1.91 × 10−8 | EPAS1, PIK3CA | Apoptosis, proliferation | [33,34] |
| miR-140-3p | −13.77 | 5.83 × 10−9 | TRIM28 | Cell growth, migration, and invasion | [35] |
| Overexpressed miRNA | Fold Change | p-value | Target | Function | Reference |
| miR-4417 | 51.35 | 1.22 × 10−12 | TGF-β; SMAD2 | Migration and mammosphere formation | [36] |
| miR-1290 | 45.78 | 1.65 × 10−12 | NAT1, IRF2 | Proliferation and invasion | [37,38] |
| miR-7641 | 45.15 | 4.43 × 10−9 | ARID1A | Proliferation | [39] |
| miR-4449 | 23.6 | 3.20 × 10−8 | SOCS3 | Proliferation | [40] |
| miR-1246 | 23.34 | 3.71 × 10−8 | CCNG2 | Migration and chemotherapy resistance | [41] |
| miR-6780b-5p | 22.19 | 5.36 × 10−8 | unknown | unknown | N/A |
| miR-3197 | 21.01 | 6.27 × 10−8 | unknown | unknown | N/A |
| miR-3175 | 20.06 | 8.10 × 10−7 | PI3K | Proliferation, invasion, and apoptosis | [42] |
| miR-1247-3p | 19.83 | 2.64 × 10−6 | unknown | unknown | N/A |
| miR-6802-5p | 18.12 | 7.17 × 10−6 | unknown | unknown | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salinas-Vera, Y.M.; Valdés, J.; Hidalgo-Miranda, A.; Cisneros-Villanueva, M.; Marchat, L.A.; Nuñez-Olvera, S.I.; Ramos-Payán, R.; Pérez-Plasencia, C.; Arriaga-Pizano, L.A.; Prieto-Chávez, J.L.; et al. Three-Dimensional Organotypic Cultures Reshape the microRNAs Transcriptional Program in Breast Cancer Cells. Cancers 2022, 14, 2490. https://doi.org/10.3390/cancers14102490
Salinas-Vera YM, Valdés J, Hidalgo-Miranda A, Cisneros-Villanueva M, Marchat LA, Nuñez-Olvera SI, Ramos-Payán R, Pérez-Plasencia C, Arriaga-Pizano LA, Prieto-Chávez JL, et al. Three-Dimensional Organotypic Cultures Reshape the microRNAs Transcriptional Program in Breast Cancer Cells. Cancers. 2022; 14(10):2490. https://doi.org/10.3390/cancers14102490
Chicago/Turabian StyleSalinas-Vera, Yarely M., Jesús Valdés, Alfredo Hidalgo-Miranda, Mireya Cisneros-Villanueva, Laurence A. Marchat, Stephanie I. Nuñez-Olvera, Rosalio Ramos-Payán, Carlos Pérez-Plasencia, Lourdes A. Arriaga-Pizano, Jessica L. Prieto-Chávez, and et al. 2022. "Three-Dimensional Organotypic Cultures Reshape the microRNAs Transcriptional Program in Breast Cancer Cells" Cancers 14, no. 10: 2490. https://doi.org/10.3390/cancers14102490
APA StyleSalinas-Vera, Y. M., Valdés, J., Hidalgo-Miranda, A., Cisneros-Villanueva, M., Marchat, L. A., Nuñez-Olvera, S. I., Ramos-Payán, R., Pérez-Plasencia, C., Arriaga-Pizano, L. A., Prieto-Chávez, J. L., & López-Camarillo, C. (2022). Three-Dimensional Organotypic Cultures Reshape the microRNAs Transcriptional Program in Breast Cancer Cells. Cancers, 14(10), 2490. https://doi.org/10.3390/cancers14102490