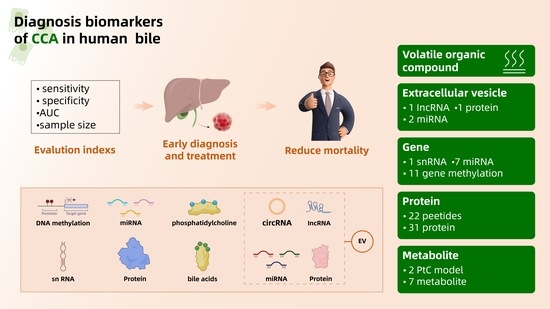
Diagnosis Biomarkers of Cholangiocarcinoma in Human Bile: An Evidence-Based Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Search for CCA Diagnosis Biomarkers in Human Bile at Gene Level
2.1. DNA Methylation and CCA Diagnosis Biomarkers
2.1.1. p16INK4a and p14ARF Promoter Methylation
2.1.2. Combined Diagnosis of Different Gene Methylation
2.1.3. Detection of DNA Methylation Biomarkers in Bile
2.2. MiRNA and CCA Diagnostic Biomarkers
2.2.1. MiR-9
2.2.2. RNU2-1f
2.2.3. Others
2.2.4. Detection of miRNA Biomarkers in Bile
3. Search for CCA Diagnosis Biomarkers in Human Bile at Protein Level
3.1. Diagnosis Biomarkers in Human Bile Proteins Based on ELISA
3.1.1. Heat Shock Proteins
3.1.2. PKM2
3.1.3. sLR11
3.1.4. sB7-H4
3.1.5. Mcm-5
3.2. Diagnosis Biomarkers in Human Bile Proteins Based on Proteomics
3.2.1. Mac-2BP
3.2.2. CEAM6
3.2.3. NGAL/MMP-9 Complex
3.2.4. AAT
3.2.5. S100A8
3.2.6. SSP411
3.2.7. TPD52 and DNAJB1
3.2.8. Others
3.3. Diagnosis Biomarkers in Human Bile Proteins Based on Glycoproteomics
3.4. Detection of Protein Biomarkers in Bile
4. Search for CCA Diagnosis Biomarkers in Human Bile at Metabolic Compounds Level
5. Search for CCA Diagnosis Biomarkers in Human Bile at the Extracellular Vesicle Level
5.1. RNA Diagnostic Biomarker of CCA in EV
5.1.1. MiRNA
5.1.2. Circ RNA
5.1.3. LncRNA
5.2. CCA Diagnostic Biomarker of Protein in EV
6. Search for CCA Diagnosis Biomarkers in Human Bile at Volatile Organic Compounds Level
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Banales, J.M.; Cardinale, V.; Carpino, G.; Marzioni, M.; Andersen, J.B.; Invernizzi, P.; Lind, G.E.; Folseraas, T.; Forbes, S.J.; Fouassier, L.; et al. Expert consensus document: Cholangiocarcinoma: Current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef] [PubMed]
- Brindley, P.J.; Bachini, M.; Ilyas, S.I.; Khan, S.A.; Loukas, A.; Sirica, A.E.; Teh, B.T.; Wongkham, S.; Gores, G.J. Cholangiocarcinoma. Nat. Rev. Dis. Primers 2021, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Bertuccio, P.; Malvezzi, M.; Carioli, G.; Hashim, D.; Boffetta, P.; El-Serag, H.B.; La Vecchia, C.; Negri, E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J. Hepatol. 2019, 71, 104–114. [Google Scholar] [CrossRef]
- Rosch, T.; Meining, A.; Fruhmorgen, S.; Zillinger, C.; Schusdziarra, V.; Hellerhoff, K.; Classen, M.; Helmberger, H. A prospective comparison of the diagnostic accuracy of ERCP, MRCP, CT, and EUS in biliary strictures. Gastrointest. Endosc. 2002, 55, 870–876. [Google Scholar] [CrossRef]
- Navaneethan, U.; Njei, B.; Lourdusamy, V.; Konjeti, R.; Vargo, J.J.; Parsi, M.A. Comparative effectiveness of biliary brush cytology and intraductal biopsy for detection of malignant biliary strictures: A systematic review and meta-analysis. Gastrointest. Endosc. 2015, 81, 168–176. [Google Scholar] [CrossRef]
- Singhi, A.D.; Nikiforova, M.N.; Chennat, J.; Papachristou, G.I.; Khalid, A.; Rabinovitz, M.; Das, R.; Sarkaria, S.; Ayasso, M.S.; Wald, A.I.; et al. Integrating next-generation sequencing to endoscopic retrograde cholangiopancreatography (ERCP)-obtained biliary specimens improves the detection and management of patients with malignant bile duct strictures. Gut 2020, 69, 52–61. [Google Scholar] [CrossRef]
- Klump, B.; Hsieh, C.J.; Dette, S.; Holzmann, K.; Kiebetalich, R.; Jung, M.; Sinn, U.; Ortner, M.; Porschen, R.; Gregor, M. Promoter methylation of INK4a/ARF as detected in bile-significance for the differential diagnosis in biliary disease. Clin. Cancer Res. 2003, 9, 1773–1778. [Google Scholar]
- Shin, S.H.; Lee, K.; Kim, B.H.; Cho, N.Y.; Jang, J.Y.; Kim, Y.T.; Kim, D.; Jang, J.J.; Kang, G.H. Bile-based detection of extrahepatic cholangiocarcinoma with quantitative DNA methylation markers and its high sensitivity. J. Mol. Diagn. 2012, 14, 256–263. [Google Scholar] [CrossRef]
- Vedeld, H.M.; Grimsrud, M.M.; Andresen, K.; Pharo, H.D.; von Seth, E.; Karlsen, T.H.; Honne, H.; Paulsen, V.; Farkkila, M.A.; Bergquist, A.; et al. Early and accurate detection of cholangiocarcinoma in patients with primary sclerosing cholangitis by methylation markers in bile. Hepatology 2022, 75, 59–73. [Google Scholar] [CrossRef]
- Navaneethan, U.; Lourdusamy, V.; Poptic, E.; Hammel, J.P.; Sanaka, M.R.; Parsi, M.A. Comparative effectiveness of pyruvate kinase M2 in bile, serum carbohydrate antigen 19-9, and biliary brushings in diagnosing malignant biliary strictures. Dig. Dis. Sci. 2015, 60, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Ayaru, L.; Stoeber, K.; Webster, G.J.; Hatfield, A.R.; Wollenschlaeger, A.; Okoturo, O.; Rashid, M.; Williams, G.; Pereira, S.P. Diagnosis of pancreaticobiliary malignancy by detection of minichromosome maintenance protein 5 in bile aspirates. Br. J. Cancer 2008, 98, 1548–1554. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Harada, K.; Sasaki, M.; Yasaka, T.; Nakanuma, Y. Heat shock proteins 27 and 70 are potential biliary markers for the detection of cholangiocarcinoma. Am. J. Pathol. 2012, 180, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Hashim Abdalla, M.S.; Taylor-Robinson, S.D.; Sharif, A.W.; Williams, H.R.; Crossey, M.M.; Badra, G.A.; Thillainayagam, A.V.; Bansi, D.S.; Thomas, H.C.; Waked, I.A.; et al. Differences in phosphatidylcholine and bile acids in bile from Egyptian and UK patients with and without cholangiocarcinoma. HPB 2011, 13, 385–390. [Google Scholar] [CrossRef]
- Navaneethan, U.; Gutierrez, N.G.; Venkatesh, P.G.; Jegadeesan, R.; Zhang, R.; Jang, S.; Sanaka, M.R.; Vargo, J.J.; Parsi, M.A.; Feldstein, A.E.; et al. Lipidomic profiling of bile in distinguishing benign from malignant biliary strictures: A single-blinded pilot study. Am. J. Gastroenterol. 2014, 109, 895–902. [Google Scholar] [CrossRef]
- Navaneethan, U.; Parsi, M.A.; Lourdusamy, V.; Bhatt, A.; Gutierrez, N.G.; Grove, D.; Sanaka, M.R.; Hammel, J.P.; Stevens, T.; Vargo, J.J.; et al. Volatile organic compounds in bile for early diagnosis of cholangiocarcinoma in patients with primary sclerosing cholangitis: A pilot study. Gastrointest. Endosc. 2015, 81, 943–949. [Google Scholar] [CrossRef]
- Metzger, J.; Negm, A.A.; Plentz, R.R.; Weismuller, T.J.; Wedemeyer, J.; Karlsen, T.H.; Dakna, M.; Mullen, W.; Mischak, H.; Manns, M.P.; et al. Urine proteomic analysis differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Gut 2013, 62, 122–130. [Google Scholar] [CrossRef]
- Levy, C.; Lymp, J.; Angulo, P.; Gores, G.J.; Larusso, N.; Lindor, K.D. The value of serum CA 19-9 in predicting cholangiocarcinomas in patients with primary sclerosing cholangitis. Dig. Dis. Sci. 2005, 50, 1734–1740. [Google Scholar] [CrossRef]
- Han, J.Y.; Ahn, K.S.; Kim, T.S.; Kim, Y.H.; Cho, K.B.; Shin, D.W.; Baek, W.K.; Suh, S.I.; Jang, B.C.; Kang, K.J. Liquid Biopsy from Bile-Circulating Tumor DNA in Patients with Biliary Tract Cancer. Cancers 2021, 13, 4581. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Ako, S.; Dohi, C.; Matsushita, H.; Matsumoto, K.; Kato, H.; Okada, H. Liquid biopsy of bile for the molecular diagnosis of gallbladder cancer. Cancer Biol. Ther. 2018, 19, 934–938. [Google Scholar] [CrossRef]
- Arechederra, M.; Rullan, M.; Amat, I.; Oyon, D.; Zabalza, L.; Elizalde, M.; Latasa, M.U.; Mercado, M.R.; Ruiz-Clavijo, D.; Saldana, C.; et al. Next-generation sequencing of bile cell-free DNA for the early detection of patients with malignant biliary strictures. Gut 2021, 71, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, A.; Demetrick, D.J.; Spillare, E.A.; Hagiwara, K.; Hussain, S.P.; Bennett, W.P.; Forrester, K.; Gerwin, B.; Serrano, M.; Beach, D.H.; et al. Mutations and altered expression of p16INK4 in human cancer. Proc. Natl. Acad. Sci. USA 1994, 91, 11045–11049. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.T.; Hasegawa, S.; Tsuda, H.; Tomioka, H.; Ushijima, M.; Noda, M.; Omura, K.; Miki, Y. Identification of a predictive gene expression signature of cervical lymph node metastasis in oral squamous cell carcinoma. Cancer Sci. 2007, 98, 740–746. [Google Scholar] [CrossRef]
- Kamp, E.; Peppelenbosch, M.P.; Doukas, M.; Verheij, J.; Ponsioen, C.Y.; van Marion, R.; Bruno, M.J.; Koerkamp, B.G.; Dinjens, W.N.M.; de Vries, A.C. Primary Sclerosing Cholangitis-Associated Cholangiocarcinoma Demonstrates High Intertumor and Intratumor Heterogeneity. Clin. Transl. Gastroenterol. 2021, 12, e00410. [Google Scholar] [CrossRef]
- Tannapfel, A.; Benicke, M.; Katalinic, A.; Uhlmann, D.; Kockerling, F.; Hauss, J.; Wittekind, C. Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut 2000, 47, 721–727. [Google Scholar] [CrossRef]
- Stott, F.J.; Bates, S.; James, M.C.; McConnell, B.B.; Starborg, M.; Brookes, S.; Palmero, I.; Ryan, K.; Hara, E.; Vousden, K.H.; et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998, 17, 5001–5014. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, B.; Du, Z.; Gao, Y.T.; Wang, Y.J.; Jing, X.; Bai, T. Identification and validation of specific methylation profile in bile for differential diagnosis of malignant biliary stricture. Clin. Biochem. 2010, 43, 1340–1344. [Google Scholar] [CrossRef]
- Randt, C.; Linschei, M. Analysis of 5-methyl-deoxycytidine in DNA by micro-HPLC. Fresenius’ Z. Für Anal. Chem. 1988, 331, 459–463. [Google Scholar] [CrossRef]
- Tost, J.; Gut, I.G. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007, 2, 2265–2275. [Google Scholar] [CrossRef]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, Y.; Xu, Q.; Ma, F.; Zhang, C.Y. Recent advances in biosensors for in vitro detection and in vivo imaging of DNA methylation. Biosens. Bioelectron. 2021, 171, 112712. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cui, Q. The relationship of human tissue microRNAs with those from body fluids. Sci. Rep. 2020, 10, 5644. [Google Scholar] [CrossRef] [PubMed]
- Cummins, J.M.; Velculescu, V.E. Implications of micro-RNA profiling for cancer diagnosis. Oncogene 2006, 25, 6220–6227. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs–microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Chen, X.; Cai, H.; Hao, X.; Li, J.; Zhang, Y.; Gao, J.; Zhou, Z.; Li, X.; Liu, C.; et al. The Emerging Roles of Autophagy-Related MicroRNAs in Cancer. Int. J. Biol. Sci. 2021, 17, 134–150. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNAs in cancer: Small molecules with a huge impact. J. Clin. Oncol. 2009, 27, 5848–5856. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Stahlhut, C.; Slack, F.J. MicroRNAs and the cancer phenotype: Profiling, signatures and clinical implications. Genome Med. 2013, 5, 111. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Xia, H.; Hui, K.M. MicroRNAs involved in regulating epithelial-mesenchymal transition and cancer stem cells as molecular targets for cancer therapeutics. Cancer Gene Ther. 2012, 19, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Y.; Li, S.H.; Huang, G.L.; Lin, G.H.; Shuang, Z.Y.; Lao, X.M.; Xu, L.; Lin, X.J.; Wang, H.Y.; Li, S.P. Identification of a novel microRNA signature associated with intrahepatic cholangiocarcinoma (ICC) patient prognosis. BMC Cancer 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. microRNA involvement in human cancer. Carcinogenesis 2012, 33, 1126–1133. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Jiao, D.; Liu, Z.; Chen, J.; Zhou, X.; Li, Z.; Li, J.; Han, X. Novel miRNA Predicts Survival and Prognosis of Cholangiocarcinoma Based on RNA-seq Data and In Vitro Experiments. Biomed. Res. Int. 2020, 2020, 5976127. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Garrido, P.; Garcia-Fernandez de Barrena, M.; Hijona, E.; Carracedo, M.; Marin, J.J.; Bujanda, L.; Banales, J.M. MicroRNAs in biliary diseases. World J. Gastroenterol. 2012, 18, 6189–6196. [Google Scholar] [CrossRef]
- Shigehara, K.; Yokomuro, S.; Ishibashi, O.; Mizuguchi, Y.; Arima, Y.; Kawahigashi, Y.; Kanda, T.; Akagi, I.; Tajiri, T.; Yoshida, H.; et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS ONE 2011, 6, e23584. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, X.; Zhang, Q.; Wang, C.; Zhao, Y. Diagnostic value of microRNAs as biomarkers for cholangiocarcinoma. Dig. Liver Dis. 2016, 48, 1227–1232. [Google Scholar] [CrossRef]
- Hildebrandt, M.A.; Gu, J.; Lin, J.; Ye, Y.; Tan, W.; Tamboli, P.; Wood, C.G.; Wu, X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 2010, 29, 5724–5728. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef]
- Tan, H.X.; Wang, Q.; Chen, L.Z.; Huang, X.H.; Chen, J.S.; Fu, X.H.; Cao, L.Q.; Chen, X.L.; Li, W.; Zhang, L.J. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med. Oncol. 2010, 27, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sanchez-Cespedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J.; et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.M.; Pu, Y.; Han, Z.; Liu, T.; Li, Y.X.; Liu, M.; Li, X.; Tang, H. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009, 276, 5537–5546. [Google Scholar] [CrossRef]
- Voigtlander, T.; Gupta, S.K.; Thum, S.; Fendrich, J.; Manns, M.P.; Lankisch, T.O.; Thum, T. MicroRNAs in Serum and Bile of Patients with Primary Sclerosing Cholangitis and/or Cholangiocarcinoma. PLoS ONE 2015, 10, e0139305. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Nopel-Dunnebacke, S.; Ahrens, M.; Jensen, S.G.; Zollner, H.; Maghnouj, A.; Wos, A.; Mayerle, J.; Munding, J.; Kost, D.; et al. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int. J. Cancer 2013, 132, E48–E57. [Google Scholar] [CrossRef]
- van Westrhenen, A.; Smidt, L.C.A.; Seute, T.; Nierkens, S.; Stork, A.C.J.; Minnema, M.C.; Snijders, T.J. Diagnostic markers for CNS lymphoma in blood and cerebrospinal fluid: A systematic review. Br. J. Haematol. 2018, 182, 384–403. [Google Scholar] [CrossRef]
- Baraniskin, A.; Zaslavska, E.; Nopel-Dunnebacke, S.; Ahle, G.; Seidel, S.; Schlegel, U.; Schmiegel, W.; Hahn, S.; Schroers, R. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for primary central nervous system lymphoma. Neuro Oncol. 2016, 18, 361–367. [Google Scholar] [CrossRef]
- Kuhlmann, J.D.; Baraniskin, A.; Hahn, S.A.; Mosel, F.; Bredemeier, M.; Wimberger, P.; Kimmig, R.; Kasimir-Bauer, S. Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer. Clin. Chem. 2014, 60, 206–213. [Google Scholar] [CrossRef]
- Kohler, J.; Schuler, M.; Gauler, T.C.; Nopel-Dunnebacke, S.; Ahrens, M.; Hoffmann, A.C.; Kasper, S.; Nensa, F.; Gomez, B.; Hahnemann, M.; et al. Circulating U2 small nuclear RNA fragments as a diagnostic and prognostic biomarker in lung cancer patients. J. Cancer Res. Clin. Oncol. 2016, 142, 795–805. [Google Scholar] [CrossRef]
- Baraniskin, A.; Nopel-Dunnebacke, S.; Schumacher, B.; Gerges, C.; Bracht, T.; Sitek, B.; Meyer, H.E.; Gerken, G.; Dechene, A.; Schlaak, J.F.; et al. Analysis of U2 small nuclear RNA fragments in the bile differentiates cholangiocarcinoma from primary sclerosing cholangitis and other benign biliary disorders. Dig. Dis. Sci. 2014, 59, 1436–1441. [Google Scholar] [CrossRef]
- Gallivanone, F.; Cava, C.; Corsi, F.; Bertoli, G.; Castiglioni, I. In Silico Approach for the Definition of radiomiRNomic Signatures for Breast Cancer Differential Diagnosis. Int. J. Mol. Sci. 2019, 20, 5825. [Google Scholar] [CrossRef] [PubMed]
- Al Rawi, N.; Elmabrouk, N.; Abu Kou, R.; Mkadmi, S.; Rizvi, Z.; Hamdoon, Z. The role of differentially expressed salivary microRNA in oral squamous cell carcinoma. A systematic review. Arch. Oral Biol. 2021, 125, 105108. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.; Kanchagar, C.; Veksler-Lublinsky, I.; Lee, R.C.; McGill, M.R.; Jaeschke, H.; Curry, S.C.; Ambros, V.R. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc. Natl. Acad. Sci. USA 2014, 111, 12169–12174. [Google Scholar] [CrossRef] [PubMed]
- Visone, R.; Rassenti, L.Z.; Veronese, A.; Taccioli, C.; Costinean, S.; Aguda, B.D.; Volinia, S.; Ferracin, M.; Palatini, J.; Balatti, V.; et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood 2009, 114, 3872–3879. [Google Scholar] [CrossRef]
- Jones, M.F.; Li, X.L.; Subramanian, M.; Shabalina, S.A.; Hara, T.; Zhu, Y.; Huang, J.; Yang, Y.; Wakefield, L.M.; Prasanth, K.V.; et al. Growth differentiation factor-15 encodes a novel microRNA 3189 that functions as a potent regulator of cell death. Cell Death Differ. 2015, 22, 1641–1653. [Google Scholar] [CrossRef]
- Han, H.S.; Kim, M.J.; Han, J.H.; Yun, J.; Kim, H.K.; Yang, Y.; Kim, K.B.; Park, S.M. Bile-derived circulating extracellular miR-30d-5p and miR-92a-3p as potential biomarkers for cholangiocarcinoma. Hepatobiliary Pancreat. Dis. Int. 2020, 19, 41–50. [Google Scholar] [CrossRef]
- Magayr, T.A.; Song, X.; Streets, A.J.; Vergoz, L.; Chang, L.; Valluru, M.K.; Yap, H.L.; Lannoy, M.; Haghighi, A.; Simms, R.J.; et al. Global microRNA profiling in human urinary exosomes reveals novel disease biomarkers and cellular pathways for autosomal dominant polycystic kidney disease. Kidney Int. 2020, 98, 420–435. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Li, W.; Ruan, K. MicroRNA detection by microarray. Anal. Bioanal. Chem. 2009, 394, 1117–1124. [Google Scholar] [CrossRef]
- Lee, J.M.; Jung, Y. Two-temperature hybridization for microarray detection of label-free microRNAs with attomole detection and superior specificity. Angew. Chem. 2011, 50, 12487–12490. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J.; Bertone, P.; Caldas, C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Masud, M.K.; Umer, M.; Hossain, M.S.A.; Yamauchi, Y.; Nguyen, N.T.; Shiddiky, M.J.A. Nanoarchitecture Frameworks for Electrochemical miRNA Detection. Trends Biochem. Sci. 2019, 44, 433–452. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat Shock Proteins and Cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef] [PubMed]
- Romani, A.A.; Crafa, P.; Desenzani, S.; Graiani, G.; Lagrasta, C.; Sianesi, M.; Soliani, P.; Borghetti, A.F. The expression of HSP27 is associated with poor clinical outcome in intrahepatic cholangiocarcinoma. BMC Cancer 2007, 7, 232. [Google Scholar] [CrossRef] [PubMed]
- Melle, C.; Ernst, G.; Escher, N.; Hartmann, D.; Schimmel, B.; Bleul, A.; Thieme, H.; Kaufmann, R.; Felix, K.; Friess, H.M.; et al. Protein profiling of microdissected pancreas carcinoma and identification of HSP27 as a potential serum marker. Clin. Chem. 2007, 53, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Prakasam, G.; Iqbal, M.A.; Bamezai, R.N.K.; Mazurek, S. Posttranslational Modifications of Pyruvate Kinase M2: Tweaks that Benefit Cancer. Front. Oncol. 2018, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Rigi, F.; Jannatabad, A.; Izanloo, A.; Roshanravan, R.; Hashemian, H.R.; Kerachian, M.A. Expression of tumor pyruvate kinase M2 isoform in plasma and stool of patients with colorectal cancer or adenomatous polyps. BMC Gastroenterol. 2020, 20, 241. [Google Scholar] [CrossRef]
- Liu, W.; Woolbright, B.L.; Pirani, K.; Didde, R.; Abbott, E.; Kaushik, G.; Martin, P.; Hamilton-Reeves, J.; Taylor, J.A., 3rd; Holzbeierlein, J.M.; et al. Tumor M2-PK: A novel urine marker of bladder cancer. PLoS ONE 2019, 14, e0218737. [Google Scholar] [CrossRef]
- Bandara, I.A.; Baltatzis, M.; Sanyal, S.; Siriwardena, A.K. Evaluation of tumor M2-pyruvate kinase (Tumor M2-PK) as a biomarker for pancreatic cancer. World J. Surg. Oncol. 2018, 16, 56. [Google Scholar] [CrossRef]
- Fujimura, K.; Ebinuma, H.; Fukamachi, I.; Ohwada, C.; Kawaguchi, T.; Shimizu, N.; Takeuchi, M.; Sakaida, E.; Jiang, M.; Nakaseko, C.; et al. Circulating LR11 is a novel soluble-receptor marker for early-stage clinical conditions in patients with non-Hodgkin’s lymphoma. Clin. Chim. Acta 2014, 430, 48–54. [Google Scholar] [CrossRef]
- Sugita, Y.; Ohwada, C.; Kawaguchi, T.; Muto, T.; Tsukamoto, S.; Takeda, Y.; Mimura, N.; Takeuchi, M.; Sakaida, E.; Shimizu, N.; et al. Prognostic impact of serum soluble LR11 in newly diagnosed diffuse large B-cell lymphoma: A multicenter prospective analysis. Clin. Chim. Acta 2016, 463, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Terai, K.; Jiang, M.; Tokuyama, W.; Murano, T.; Takada, N.; Fujimura, K.; Ebinuma, H.; Kishimoto, T.; Hiruta, N.; Schneider, W.J.; et al. Levels of soluble LR11/SorLA are highly increased in the bile of patients with biliary tract and pancreatic cancers. Clin. Chim. Acta 2016, 457, 130–136. [Google Scholar] [CrossRef]
- MacGregor, H.L.; Ohashi, P.S. Molecular Pathways: Evaluating the Potential for B7-H4 as an Immunoregulatory Target. Clin. Cancer Res. 2017, 23, 2934–2941. [Google Scholar] [CrossRef] [PubMed]
- Ke, W.; Zeng, L.; Hu, Y.; Chen, S.; Tian, M.; Hu, Q. Detection of early-stage extrahepatic cholangiocarcinoma in patients with biliary strictures by soluble B7-H4 in the bile. Am. J. Cancer Res. 2018, 8, 699–707. [Google Scholar] [PubMed]
- Semple, J.W.; Duncker, B.P. ORC-associated replication factors as biomarkers for cancer. Biotechnol. Adv. 2004, 22, 621–631. [Google Scholar] [CrossRef]
- Stoeber, K.; Swinn, R.; Prevost, A.T.; de Clive-Lowe, P.; Halsall, I.; Dilworth, S.M.; Marr, J.; Turner, W.H.; Bullock, N.; Doble, A.; et al. Diagnosis of genito-urinary tract cancer by detection of minichromosome maintenance 5 protein in urine sediments. J. Natl. Cancer Inst. 2002, 94, 1071–1079. [Google Scholar] [CrossRef]
- Williams, G.H.; Swinn, R.; Prevost, A.T.; De Clive-Lowe, P.; Halsall, I.; Going, J.J.; Hales, C.N.; Stoeber, K.; Middleton, S.J. Diagnosis of oesophageal cancer by detection of minichromosome maintenance 5 protein in gastric aspirates. Br. J. Cancer 2004, 91, 714–719. [Google Scholar] [CrossRef]
- Pandey, A.; Mann, M. Proteomics to study genes and genomes. Nature 2000, 405, 837–846. [Google Scholar] [CrossRef]
- Clarke, W.; Zhang, Z.; Chan, D.W. The application of clinical proteomics to cancer and other diseases. Clin. Chem. Lab. Med. 2003, 41, 1562–1570. [Google Scholar] [CrossRef]
- Wulfkuhle, J.D.; Liotta, L.A.; Petricoin, E.F. Proteomic applications for the early detection of cancer. Nat. Rev. Cancer 2003, 3, 267–275. [Google Scholar] [CrossRef]
- Lankisch, T.O.; Metzger, J.; Negm, A.A.; Vosskuhl, K.; Schiffer, E.; Siwy, J.; Weismuller, T.J.; Schneider, A.S.; Thedieck, K.; Baumeister, R.; et al. Bile proteomic profiles differentiate cholangiocarcinoma from primary sclerosing cholangitis and choledocholithiasis. Hepatology 2011, 53, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Brakebusch, C.; Engel, J.; Timpl, R. Mac-2 binding protein is a cell-adhesive protein of the extracellular matrix which self-assembles into ring-like structures and binds beta1 integrins, collagens and fibronectin. EMBO J. 1998, 17, 1606–1613. [Google Scholar] [CrossRef]
- Kamada, Y.; Ono, M.; Hyogo, H.; Fujii, H.; Sumida, Y.; Yamada, M.; Mori, K.; Tanaka, S.; Maekawa, T.; Ebisutani, Y.; et al. Use of Mac-2 binding protein as a biomarker for nonalcoholic fatty liver disease diagnosis. Hepatol. Commun. 2017, 1, 780–791. [Google Scholar] [CrossRef]
- Sardana, G.; Marshall, J.; Diamandis, E.P. Discovery of candidate tumor markers for prostate cancer via proteomic analysis of cell culture-conditioned medium. Clin. Chem. 2007, 53, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Koopmann, J.; Thuluvath, P.J.; Zahurak, M.L.; Kristiansen, T.Z.; Pandey, A.; Schulick, R.; Argani, P.; Hidalgo, M.; Iacobelli, S.; Goggins, M.; et al. Mac-2-binding protein is a diagnostic marker for biliary tract carcinoma. Cancer 2004, 101, 1609–1615. [Google Scholar] [CrossRef] [PubMed]
- Rizeq, B.; Zakaria, Z.; Ouhtit, A. Towards understanding the mechanisms of actions of carcinoembryonic antigen-related cell adhesion molecule 6 in cancer progression. Cancer Sci. 2018, 109, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Farina, A.; Dumonceau, J.M.; Antinori, P.; Annessi-Ramseyer, I.; Frossard, J.L.; Hochstrasser, D.F.; Delhaye, M.; Lescuyer, P. Bile carcinoembryonic cell adhesion molecule 6 (CEAM6) as a biomarker of malignant biliary stenoses. Biochim. Biophys. Acta BBA-Proteins Proteom. 2014, 1844, 1018–1025. [Google Scholar] [CrossRef]
- Lukic, N.; Visentin, R.; Delhaye, M.; Frossard, J.L.; Lescuyer, P.; Dumonceau, J.M.; Farina, A. An integrated approach for comparative proteomic analysis of human bile reveals overexpressed cancer-associated proteins in malignant biliary stenosis. Biochim. Biophys. Acta BBA-Proteins Proteom. 2014, 1844, 1026–1033. [Google Scholar] [CrossRef]
- Rose, J.B.; Correa-Gallego, C.; Li, Y.; Nelson, J.; Alseidi, A.; Helton, W.S.; Allen, P.J.; D’Angelica, M.I.; DeMatteo, R.P.; Fong, Y.; et al. The Role of Biliary Carcinoembryonic Antigen-Related Cellular Adhesion Molecule 6 (CEACAM6) as a Biomarker in Cholangiocarcinoma. PLoS ONE 2016, 11, e0150195. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kaur, S.; Guha, S.; Batra, S.K. The multifaceted roles of neutrophil gelatinase associated lipocalin (NGAL) in inflammation and cancer. Biochim. Biophys. Acta BBA-Rev. Cancer 2012, 1826, 129–169. [Google Scholar] [CrossRef]
- Zabron, A.A.; Horneffer-van der Sluis, V.M.; Wadsworth, C.A.; Laird, F.; Gierula, M.; Thillainayagam, A.V.; Vlavianos, P.; Westaby, D.; Taylor-Robinson, S.D.; Edwards, R.J.; et al. Elevated levels of neutrophil gelatinase-associated lipocalin in bile from patients with malignant pancreatobiliary disease. Am. J. Gastroenterol. 2011, 106, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Budzynska, A.; Nowakowska-Dulawa, E.; Marek, T.; Boldys, H.; Nowak, A.; Hartleb, M. Differentiation of pancreatobiliary cancer from benign biliary strictures using neutrophil gelatinase-associated lipocalin. J. Physiol. Pharmacol. 2013, 64, 109–114. [Google Scholar] [PubMed]
- Chen, L.; Zhang, J.; He, Y.; Ding, X.Y. Matrix metalloproteinase-9 expression of GCTSC in peripheral tissue and central tissue of GCTB. J. Cell. Biochem. 2018, 119, 5805–5812. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Prieto, S.; Barcia-Castro, L.; Paez de la Cadena, M.; Rodriguez-Berrocal, F.J.; Vazquez-Iglesias, L.; Botana-Rial, M.I.; Fernandez-Villar, A.; De Chiara, L. Relevance of matrix metalloproteases in non-small cell lung cancer diagnosis. BMC Cancer 2017, 17, 823. [Google Scholar] [CrossRef] [PubMed]
- Lubowicka, E.; Gacuta, E.; Zajkowska, M.; Głażewska, E.K.; Przylipiak, A.; Chrostek, L.; Zbucka-Krętowska, M.; Ławicki, S. [The plasma levels and diagnostic utility of matrix metalloproteinase-9 and CA 125 in cervical cancer patients]. Pol. Merkur. Lek. Organ Pol. Tow. Lek. 2017, 43, 10–14. [Google Scholar]
- Li, L.N.; Zhou, X.; Gu, Y.; Yan, J. Prognostic value of MMP-9 in ovarian cancer: A meta-analysis. Asian Pac. J. Cancer Prev. 2013, 14, 4107–4113. [Google Scholar] [CrossRef]
- Tian, M.; Cui, Y.Z.; Song, G.H.; Zong, M.J.; Zhou, X.Y.; Chen, Y.; Han, J.X. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer 2008, 8, 241. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Q.; Yuan, T.X.; Song, Q.L.; Zhang, Y.; Wei, Q.; Zhou, L.; Luo, J.; Zuo, G.; Tang, M.; et al. Matrix metalloproteinase 9 (MMP-9) in osteosarcoma: Review and meta-analysis. Clin. Chim. Acta 2014, 433, 225–231. [Google Scholar] [CrossRef]
- Yousef, E.M.; Tahir, M.R.; St-Pierre, Y.; Gaboury, L.A. MMP-9 expression varies according to molecular subtypes of breast cancer. BMC Cancer 2014, 14, 609. [Google Scholar] [CrossRef]
- Okada, N.; Ishida, H.; Murata, N.; Hashimoto, D.; Seyama, Y.; Kubota, S. Matrix metalloproteinase-2 and -9 in bile as a marker of liver metastasis in colorectal cancer. Biochem. Biophys. Res. Commun. 2001, 288, 212–216. [Google Scholar] [CrossRef]
- Ince, A.T.; Yildiz, K.; Gangarapu, V.; Kayar, Y.; Baysal, B.; Karatepe, O.; Kemik, A.S.; Senturk, H. Serum and biliary MMP-9 and TIMP-1 concentrations in the diagnosis of cholangiocarcinoma. Int. J. Clin. Exp. Med. 2015, 8, 2734–2740. [Google Scholar] [PubMed]
- Fernandez, C.A.; Yan, L.; Louis, G.; Yang, J.; Kutok, J.L.; Moses, M.A. The matrix metalloproteinase-9/neutrophil gelatinase-associated lipocalin complex plays a role in breast tumor growth and is present in the urine of breast cancer patients. Clin. Cancer Res. 2005, 11, 5390–5395. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.F.; Hu, Y.Y.; Jin, T.; Xu, K.; Wang, S.H.; Du, G.Z.; Wu, B.L.; Li, L.Y.; Xu, L.Y.; Li, E.M.; et al. Matrix Metalloproteinase-9/Neutrophil Gelatinase-Associated Lipocalin Complex Activity in Human Glioma Samples Predicts Tumor Presence and Clinical Prognosis. Dis. Markers 2015, 2015, 138974. [Google Scholar] [CrossRef] [PubMed]
- Shimura, T.; Dagher, A.; Sachdev, M.; Ebi, M.; Yamada, T.; Yamada, T.; Joh, T.; Moses, M.A. Urinary ADAM12 and MMP-9/NGAL complex detect the presence of gastric cancer. Cancer Prev. Res. 2015, 8, 240–248. [Google Scholar] [CrossRef]
- Guette, C.; Valo, I.; Vetillard, A.; Coqueret, O. Olfactomedin-4 is a candidate biomarker of solid gastric, colorectal, pancreatic, head and neck, and prostate cancers. Proteom. Clin. Appl. 2015, 9, 58–63. [Google Scholar] [CrossRef]
- Lou, S.; Wang, P.; Yang, J.; Ma, J.; Liu, C.; Zhou, M. Prognostic and Clinicopathological Value of Rac1 in Cancer Survival: Evidence from a Meta-Analysis. J. Cancer 2018, 9, 2571–2579. [Google Scholar] [CrossRef]
- Sun, Z.; Yang, P. Role of imbalance between neutrophil elastase and alpha 1-antitrypsin in cancer development and progression. Lancet Oncol. 2004, 5, 182–190. [Google Scholar] [CrossRef]
- Laohaviroj, M.; Potriquet, J.; Jia, X.; Suttiprapa, S.; Chamgramol, Y.; Pairojkul, C.; Sithithaworn, P.; Mulvenna, J.; Sripa, B. A comparative proteomic analysis of bile for biomarkers of cholangiocarcinoma. Tumour Biol. 2017, 39, 1010428317705764. [Google Scholar] [CrossRef]
- Son, K.H.; Ahn, C.B.; Kim, H.J.; Kim, J.S. Quantitative proteomic analysis of bile in extrahepatic cholangiocarcinoma patients. J. Cancer 2020, 11, 4073–4080. [Google Scholar] [CrossRef]
- Jamnongkan, W.; Techasen, A.; Thanan, R.; Duenngai, K.; Sithithaworn, P.; Mairiang, E.; Loilome, W.; Namwat, N.; Pairojkul, C.; Yongvanit, P. Oxidized alpha-1 antitrypsin as a predictive risk marker of opisthorchiasis-associated cholangiocarcinoma. Tumour Biol. 2013, 34, 695–704. [Google Scholar] [CrossRef]
- Jukic, A.; Bakiri, L.; Wagner, E.F.; Tilg, H.; Adolph, T.E. Calprotectin: From biomarker to biological function. Gut 2021, 70, 1978–1988. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, Y.; Yamazaki, M.; Chazin, W.J.; Yui, S. Regulation of S100A8/A9 (calprotectin) binding to tumor cells by zinc ion and its implication for apoptosis-inducing activity. Mediat. Inflamm. 2005, 2005, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ai, K.X.; Yuan, Z.; Huang, X.Y.; Zhang, H.Z. Different expression of S100A8 in malignant and benign gallbladder diseases. Dig. Dis. Sci. 2013, 58, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Hu, Y.; Hu, M.; Xu, Y.; Chen, M.; Du, C.; Cui, J.; Zheng, P.; Lai, J.; Zhang, Y.; et al. [Corrigendum] S100A8 facilitates cholangiocarcinoma metastasis via upregulation of VEGF through TLR4/NFkappaB pathway activation. Int. J. Oncol. 2020, 56, 1046. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Fan, W.; Liu, J.; Huang, B.; Cheng, Q.; Wang, P.; Duan, Y.; Ma, T.; Chen, L.; Wang, Y.; et al. Prognostic Role of S100A8 in Human Solid Cancers: A Systematic Review and Validation. Front. Oncol. 2020, 10, 564248. [Google Scholar] [CrossRef]
- Yasar, O.; Akcay, T.; Obek, C.; Turegun, F.A. Significance of S100A8, S100A9 and calprotectin levels in bladder cancer. Scand. J. Clin. Lab. Investig. 2017, 77, 437–441. [Google Scholar] [CrossRef]
- Duangkumpha, K.; Stoll, T.; Phetcharaburanin, J.; Yongvanit, P.; Thanan, R.; Techasen, A.; Namwat, N.; Khuntikeo, N.; Chamadol, N.; Roytrakul, S.; et al. Discovery and Qualification of Serum Protein Biomarker Candidates for Cholangiocarcinoma Diagnosis. J. Proteome Res. 2019, 18, 3305–3316. [Google Scholar] [CrossRef]
- Shi, Y.; Deng, X.; Zhan, Q.; Shen, B.; Jin, X.; Zhu, Z.; Chen, H.; Li, H.; Peng, C. A prospective proteomic-based study for identifying potential biomarkers for the diagnosis of cholangiocarcinoma. J. Gastrointest. Surg. 2013, 17, 1584–1591. [Google Scholar] [CrossRef]
- Wu, W.; Juan, W.C.; Liang, C.R.; Yeoh, K.G.; So, J.; Chung, M.C. S100A9, GIF and AAT as potential combinatorial biomarkers in gastric cancer diagnosis and prognosis. Proteom. Clin. Appl. 2012, 6, 152–162. [Google Scholar] [CrossRef]
- Meng, J.; Gu, F.; Fang, H.; Qu, B. Elevated Serum S100A9 Indicated Poor Prognosis in Hepatocellular Carcinoma after Curative Resection. J. Cancer 2019, 10, 408–415. [Google Scholar] [CrossRef]
- Hermani, A.; Hess, J.; De Servi, B.; Medunjanin, S.; Grobholz, R.; Trojan, L.; Angel, P.; Mayer, D. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin. Cancer Res. 2005, 11, 5146–5152. [Google Scholar] [CrossRef] [PubMed]
- Moris, D.; Spartalis, E.; Angelou, A.; Margonis, G.A.; Papalambros, A.; Petrou, A.; Athanasiou, A.; Schizas, D.; Dimitroulis, D.; Felekouras, E. The value of calprotectin S100A8/A9 complex as a biomarker in colorectal cancer: A systematic review. J. BU ON 2016, 21, 859–866. [Google Scholar]
- Şumnu, Ş.; Mehtap, Ö.; Mersin, S.; Toptaş, T.; Görür, G.; Gedük, A.; Ünal, S.; Polat, M.G.; Aygün, K.; Yenihayat, E.M.; et al. Serum calprotectin (S100A8/A9) levels as a new potential biomarker of treatment response in Hodgkin lymphoma. Int. J. Lab. Hematol. 2021, 43, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.J.; Wu, A.Z.; Santos, M.; Feng, Z.M.; Huang, L.; Chen, Y.M.; Zhu, K.; Chen, C.L. Cloning and characterization of rat spermatid protein SSP411: A thioredoxin-like protein. J. Androl. 2004, 25, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, W.; Wu, J.; Feng, B.; Chen, W.; Wang, M.; Tang, J.; Wang, F.; Cheng, F.; Pu, L.; et al. Comparative proteomic profiling of human bile reveals SSP411 as a novel biomarker of cholangiocarcinoma. PLoS ONE 2012, 7, e47476. [Google Scholar] [CrossRef]
- Zhong, A.; Chen, T.; Zhou, T.; Zhang, Z.; Shi, M. TPD52L2 Is a Prognostic Biomarker and Correlated With Immune Infiltration in Lung Adenocarcinoma. Front. Pharmacol. 2021, 12, 728420. [Google Scholar] [CrossRef]
- Kong, L.; Liu, P.; Fei, X.; Wu, T.; Wang, Z.; Zhang, B.; Li, J.; Tan, X. A Prognostic Prediction Model Developed Based on Four CpG Sites and Weighted Correlation Network Analysis Identified DNAJB1 as a Novel Biomarker for Pancreatic Cancer. Front. Oncol. 2020, 10, 1716. [Google Scholar] [CrossRef]
- Ren, H.; Luo, M.; Chen, J.; Zhou, Y.; Li, X.; Zhan, Y.; Shen, D.; Chen, B. Identification of TPD52 and DNAJB1 as two novel bile biomarkers for cholangiocarcinoma by iTRAQbased quantitative proteomics analysis. Oncol. Rep. 2019, 42, 2622–2634. [Google Scholar] [CrossRef]
- Sripa, B.; Bethony, J.M.; Sithithaworn, P.; Kaewkes, S.; Mairiang, E.; Loukas, A.; Mulvenna, J.; Laha, T.; Hotez, P.J.; Brindley, P.J. Opisthorchiasis and Opisthorchis-associated cholangiocarcinoma in Thailand and Laos. Acta Trop. 2011, 120 (Suppl. S1), S158–S168. [Google Scholar] [CrossRef]
- Aksorn, N.; Roytrakul, S.; Kittisenachai, S.; Leelawat, K.; Chanvorachote, P.; Topanurak, S.; Hamano, S.; Lek-Uthai, U. Novel Potential Biomarkers for Opisthorchis viverrini Infection and Associated Cholangiocarcinoma. In Vivo 2018, 32, 871–878. [Google Scholar] [CrossRef]
- Voigtlander, T.; Metzger, J.; Schonemeier, B.; Jager, M.; Mischak, H.; Manns, M.P.; Lankisch, T.O. A combined bile and urine proteomic test for cholangiocarcinoma diagnosis in patients with biliary strictures of unknown origin. United Eur. Gastroenterol. J. 2017, 5, 668–676. [Google Scholar] [CrossRef]
- Voigtlander, T.; Metzger, J.; Husi, H.; Kirstein, M.M.; Pejchinovski, M.; Latosinska, A.; Frantzi, M.; Mullen, W.; Book, T.; Mischak, H.; et al. Bile and urine peptide marker profiles: Access keys to molecular pathways and biological processes in cholangiocarcinoma. J. Biomed. Sci. 2020, 27, 13. [Google Scholar] [CrossRef]
- Esteller, A. Physiology of bile secretion. World J. Gastroenterol. 2008, 14, 5641–5649. [Google Scholar] [CrossRef]
- Ciordia, S.; Alvarez-Sola, G.; Rullan, M.; Urman, J.M.; Avila, M.A.; Corrales, F.J. Bile Processing Protocol for Improved Proteomic Analysis. Methods Mol. Biol. 2022, 2420, 1–10. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Kawamoto, T.; Matsuzaki, H.; Irimura, T.; Ikehara, Y.; Zen, Y.; Nakanuma, Y.; Yamamoto, M.; Ohkohchi, N.; et al. Wisteria floribunda agglutinin-positive mucin 1 is a sensitive biliary marker for human cholangiocarcinoma. Hepatology 2010, 52, 174–182. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Matsuzaki, H.; Kawamoto, T.; Shikanai, T.; Nakanuma, Y.; Yamamoto, M.; Ohkohchi, N.; Ikehara, Y.; Shoda, J.; et al. Glycoproteomics-based cancer marker discovery adopting dual enrichment with Wisteria floribunda agglutinin for high specific glyco-diagnosis of cholangiocarcinoma. J. Proteom. 2013, 85, 1–11. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Yokoyama, Y.; Ebata, T.; Matsuda, A.; Kuno, A.; Ikehara, Y.; Shoda, J.; Narimatsu, H.; Nagino, M. Verification of WFA-Sialylated MUC1 as a Sensitive Biliary Biomarker for Human Biliary Tract Cancer. Ann. Surg. Oncol. 2016, 23, 671–677. [Google Scholar] [CrossRef]
- Wongkham, S.; Sheehan, J.K.; Boonla, C.; Patrakitkomjorn, S.; Howard, M.; Kirkham, S.; Sripa, B.; Wongkham, C.; Bhudhisawasdi, V. Serum MUC5AC mucin as a potential marker for cholangiocarcinoma. Cancer Lett. 2003, 195, 93–99. [Google Scholar] [CrossRef]
- Shibahara, H.; Tamada, S.; Higashi, M.; Goto, M.; Batra, S.K.; Hollingsworth, M.A.; Imai, K.; Yonezawa, S. MUC4 is a novel prognostic factor of intrahepatic cholangiocarcinoma-mass forming type. Hepatology 2004, 39, 220–229. [Google Scholar] [CrossRef]
- Tamada, S.; Shibahara, H.; Higashi, M.; Goto, M.; Batra, S.K.; Imai, K.; Yonezawa, S. MUC4 is a novel prognostic factor of extrahepatic bile duct carcinoma. Clin. Cancer Res. 2006, 12, 4257–4264. [Google Scholar] [CrossRef]
- Matull, W.R.; Andreola, F.; Loh, A.; Adiguzel, Z.; Deheragoda, M.; Qureshi, U.; Batra, S.K.; Swallow, D.M.; Pereira, S.P. MUC4 and MUC5AC are highly specific tumour-associated mucins in biliary tract cancer. Br. J. Cancer 2008, 98, 1675–1681. [Google Scholar] [CrossRef] [PubMed]
- Hanash, S.M.; Pitteri, S.J.; Faca, V.M. Mining the plasma proteome for cancer biomarkers. Nature 2008, 452, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Selegard, R.; Aili, D.; Liedberg, B. Peptide functionalized gold nanoparticles for colorimetric detection of matrilysin (MMP-7) activity. Nanoscale 2013, 5, 8973–8976. [Google Scholar] [CrossRef]
- Xia, Z.; Xing, Y.; So, M.K.; Koh, A.L.; Sinclair, R.; Rao, J. Multiplex detection of protease activity with quantum dot nanosensors prepared by intein-mediated specific bioconjugation. Anal. Chem. 2008, 80, 8649–8655. [Google Scholar] [CrossRef]
- Kou, B.B.; Zhang, L.; Xie, H.; Wang, D.; Yuan, Y.L.; Chai, Y.Q.; Yuan, R. DNA Enzyme-Decorated DNA Nanoladders as Enhancer for Peptide Cleavage-Based Electrochemical Biosensor. ACS Appl. Mater. Interfaces 2016, 8, 22869–22874. [Google Scholar] [CrossRef] [PubMed]
- Beyoglu, D.; Idle, J.R. The metabolomic window into hepatobiliary disease. J. Hepatol. 2013, 59, 842–858. [Google Scholar] [CrossRef] [PubMed]
- Sharif, A.W.; Williams, H.R.; Lampejo, T.; Khan, S.A.; Bansi, D.S.; Westaby, D.; Thillainayagam, A.V.; Thomas, H.C.; Cox, I.J.; Taylor-Robinson, S.D. Metabolic profiling of bile in cholangiocarcinoma using in vitro magnetic resonance spectroscopy. HPB 2010, 12, 396–402. [Google Scholar] [CrossRef]
- Albiin, N.; Smith, I.C.; Arnelo, U.; Lindberg, B.; Bergquist, A.; Dolenko, B.; Bryksina, N.; Bezabeh, T. Detection of cholangiocarcinoma with magnetic resonance spectroscopy of bile in patients with and without primary sclerosing cholangitis. Acta Radiol. 2008, 49, 855–862. [Google Scholar] [CrossRef]
- Park, J.Y.; Park, B.K.; Ko, J.S.; Bang, S.; Song, S.Y.; Chung, J.B. Bile acid analysis in biliary tract cancer. Yonsei Med. J. 2006, 47, 817–825. [Google Scholar] [CrossRef]
- Song, W.S.; Park, H.M.; Ha, J.M.; Shin, S.G.; Park, H.G.; Kim, J.; Zhang, T.; Ahn, D.H.; Kim, S.M.; Yang, Y.H.; et al. Discovery of glycocholic acid and taurochenodeoxycholic acid as phenotypic biomarkers in cholangiocarcinoma. Sci. Rep. 2018, 8, 11088. [Google Scholar] [CrossRef]
- Khan, S.A.; Cox, I.J.; Thillainayagam, A.V.; Bansi, D.S.; Thomas, H.C.; Taylor-Robinson, S.D. Proton and phosphorus-31 nuclear magnetic resonance spectroscopy of human bile in hepatopancreaticobiliary cancer. Eur. J. Gastroenterol. Hepatol. 2005, 17, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Volinsky, R.; Kinnunen, P.K. Oxidized phosphatidylcholines in membrane-level cellular signaling: From biophysics to physiology and molecular pathology. FEBS J. 2013, 280, 2806–2816. [Google Scholar] [CrossRef] [PubMed]
- Gómez, C.; Stücheli, S.; Kratschmar, D.V.; Bouitbir, J.; Odermatt, A. Development and Validation of a Highly Sensitive LC-MS/MS Method for the Analysis of Bile Acids in Serum, Plasma, and Liver Tissue Samples. Metabolites 2020, 10, 282. [Google Scholar] [CrossRef] [PubMed]
- Roda, A.; Girotti, S.; Ghini, S.; Grigolo, B.; Carrea, G.; Bovara, R. Continuous-flow determination of primary bile acids, by bioluminescence, with use of nylon-immobilized bacterial enzymes. Clin. Chem. 1984, 30, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Prescot, A.P.; Collins, D.J.; Leach, M.O.; Dzik-Jurasz, A.S. Human gallbladder bile: Noninvasive investigation in vivo with single-voxel 1H MR spectroscopy. Radiology 2003, 229, 587–592. [Google Scholar] [CrossRef]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Aguero, R.; Lacasta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef]
- Severino, V.; Dumonceau, J.M.; Delhaye, M.; Moll, S.; Annessi-Ramseyer, I.; Robin, X.; Frossard, J.L.; Farina, A. Extracellular Vesicles in Bile as Markers of Malignant Biliary Stenoses. Gastroenterology 2017, 153, 495–504. [Google Scholar] [CrossRef]
- Han, J.Y.; Ahn, K.S.; Kim, Y.H.; Kim, T.S.; Baek, W.K.; Suh, S.I.; Kang, K.J. Circulating microRNAs as biomarkers in bile-derived exosomes of cholangiocarcinoma. Ann. Surg. Treat. Res. 2021, 101, 140–150. [Google Scholar] [CrossRef]
- Gao, L.; Yang, X.; Zhang, H.; Yu, M.; Long, J.; Yang, T. Inhibition of miR-10a-5p suppresses cholangiocarcinoma cell growth through downregulation of Akt pathway. Onco Targets Ther. 2018, 11, 6981–6994. [Google Scholar] [CrossRef]
- Li, L.; Masica, D.; Ishida, M.; Tomuleasa, C.; Umegaki, S.; Kalloo, A.N.; Georgiades, C.; Singh, V.K.; Khashab, M.; Amateau, S.; et al. Human bile contains microRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology 2014, 60, 896–907. [Google Scholar] [CrossRef]
- Xu, Y.; Leng, K.; Yao, Y.; Kang, P.; Liao, G.; Han, Y.; Shi, G.; Ji, D.; Huang, P.; Zheng, W.; et al. A Circular RNA, Cholangiocarcinoma-Associated Circular RNA 1, Contributes to Cholangiocarcinoma Progression, Induces Angiogenesis, and Disrupts Vascular Endothelial Barriers. Hepatology 2021, 73, 1419–1435. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, L.; Li, Z.; Zhong, X.; Tai, S.; Jiang, X.; Cui, Y. Functions and roles of long noncoding RNA in cholangiocarcinoma. J. Cell. Physiol. 2019, 234, 17113–17126. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Wang, Y.; Nie, J.; Li, Q.; Tang, L.; Deng, X.; Wang, F.; Xu, B.; Wu, X.; Zhang, X.; et al. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget 2017, 8, 69995–70005. [Google Scholar] [CrossRef] [PubMed]
- Hurd, T.W.; Gao, L.; Roh, M.H.; Macara, I.G.; Margolis, B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 2003, 5, 137–142. [Google Scholar] [CrossRef]
- Ikeda, C.; Haga, H.; Makino, N.; Inuzuka, T.; Kurimoto, A.; Ueda, T.; Matsuda, A.; Kakizaki, Y.; Ishizawa, T.; Kobayashi, T.; et al. Utility of Claudin-3 in extracellular vesicles from human bile as biomarkers of cholangiocarcinoma. Sci. Rep. 2021, 11, 1195. [Google Scholar] [CrossRef]
- Navaneethan, U.; Parsi, M.A.; Gutierrez, N.G.; Bhatt, A.; Venkatesh, P.G.; Lourdusamy, D.; Grove, D.; Hammel, J.P.; Jang, S.; Sanaka, M.R.; et al. Volatile organic compounds in bile can diagnose malignant biliary strictures in the setting of pancreatic cancer: A preliminary observation. Gastrointest. Endosc. 2014, 80, 1038–1045. [Google Scholar] [CrossRef]
- Rupp, C.; Bode, K.A.; Leopold, Y.; Sauer, P.; Gotthardt, D.N. Pathological features of primary sclerosing cholangitis identified by bile proteomic analysis. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2018, 1864, 1380–1389. [Google Scholar] [CrossRef]


| Type | Name | Sensitivity | Specificity | AUC | SS | Ref | Remark |
|---|---|---|---|---|---|---|---|
| Gene | p16INK4a↑ | 53.6% | 93.8% | \ | 7 | [8] | |
| p14ARF↑ | 46.2% | 96.9% | \ | 7 | [8] | ||
| CCND2↑ | 75.6% | 100.0% | \ | 125 | [9] | CD | |
| CDH13↑ | 75.6% | 100.0% | \ | 125 | [9] | CD | |
| GRIN2B↑ | 75.6% | 100.0% | \ | 125 | [9] | CD | |
| RUNX3↑ | 75.6% | 100.0% | \ | 125 | [9] | CD | |
| TWIST1↑ | 75.6% | 100.0% | \ | 125 | [9] | CD | |
| CDO1↑ | 100.0% | 90.0% | \ | 344 | [10] | CD | |
| CNRIP1↑ | 100.0% | 90.0% | \ | 344 | [10] | CD | |
| SEPT9↑ | 100.0% | 90.0% | \ | 344 | [10] | CD | |
| VIM↑ | 100.0% | 90.0% | \ | 344 | [10] | CD | |
| miR-9↑ | 88.9% | 100.0% | 97.5% | 18 | [46] | ||
| miR-1537↑ | 67.0% | 90.0% | 78.0% | 83 | [54] | ||
| miR-640↑ | 50.0% | 92.0% | 81.0% | 83 | [54] | ||
| miR-3189↑ | 67.0% | 89.0% | 80.0% | 83 | [54] | ||
| miR-412↑ | 50.0% | 89.0% | 81.0% | 83 | [54] | ||
| RNU2-1f↑ | 67.0% | 91.0% | 85.6% | 34 | [60] | ||
| miR-30d-5p↑ | 81.1% | 60.5% | 73.0% | 106 | [66] | ||
| miR-92a-3p↑ | 65.7% | 66.7% | 65.2% | 106 | [66] | ||
| Protein | HSP27↑ | 90.0% | 90.0% | 86.0% | 20 | [13] | |
| HSP70↑ | 80.0% | 80.0% | 80.5% | 20 | [13] | ||
| PKM2↑ | 52.9% | 94.1% | 77.0% | 74 | [11] | ||
| sLR11↑ | 100.0% | 80.0% | 89.0% | 147 | [82] | ||
| B7-H3↑ | 81.7% | 69.1% | 83.7% | 213 | [84] | ||
| B7-H3 + ETFB | 88.1% | 100.0% | 93.9% | 213 | [84] | CD | |
| Mcm-5↑ | 66.0% | 94.0% | 80.0% | 106 | [12] | ||
| Mac-2BP↑ | 69.0% | 67.0% | 70.0% | 78 | [95] | ||
| Mac-2BP + serum CA19-9 | \ | \ | 75.0% | 78 | [95] | CD | |
| CEAM6↑ | 83.0% | 93.0% | 92.0% | 41 | [97] | ||
| CEAM6 + serum CA19-9 | \ | \ | 96.0% | 41 | [97] | CD | |
| CEAM6↑ | 87.5% | 69.1% | 74.0% | 83 | [98] | ||
| NGAL↑ | 94.0% | 55.0% | 76.0% | 59 | [101] | ||
| NGAL↑ | 77.3% | 72.2% | 74.0% | 40 | [102] | ||
| MMP-9↑ | 93.9% | 32.5% | 50.4% | 113 | [111] | ||
| TIMP-1↓ | 96.9% | 36.2% | 53.9% | 113 | [111] | ||
| OLFM4↑ | \ | \ | \ | 13 | [98] | ||
| SDCB2↑ | \ | \ | \ | 13 | [98] | ||
| RAC1↑ | \ | \ | \ | 13 | [98] | ||
| AAT↑ | 70.0% | \ | \ | 54 | [118] | ||
| S100A8↑ | \ | \ | \ | 8 | [123] | ||
| SSP411↑ | \ | \ | \ | 25 | [135] | ||
| PGAM1↑ | \ | \ | \ | 25 | [135] | ||
| PDIA3↑ | \ | \ | \ | 25 | [135] | ||
| HSPD1↑ | \ | \ | \ | 25 | [135] | ||
| APOM↓ | \ | \ | \ | 25 | [135] | ||
| TPD52↑ | \ | \ | \ | 37 | [138] | ||
| DNAJB1↑ | \ | \ | \ | 37 | [138] | ||
| immunoglobulin heavy chain↑ | \ | \ | \ | 43 | [140] | ||
| TLS↑ | \ | \ | \ | 43 | [140] | ||
| VSX2↑ | \ | \ | \ | 43 | [140] | ||
| 22 peptides model | \ | \ | 100.0% | 107 | [91] | ||
| MUC1↑ | 90.0% | 72.0% | 85.0% | 58 | [145] | ||
| L1CAM↑ | 66.0% | 93.0% | 82.0% | 58 | [145] | ||
| MUC1 + L1CAM | 90.0% | 79.0% | 93.0% | 58 | [145] | CD | |
| MUC4↑ | 27.0% | 99.0% | \ | 62 | [151] | ||
| MUC4 + serum MUC5AC | 58.0% | 87.0% | \ | 61 | [151] | CD | |
| Metabolite | GCA↑ | \ | \ | \ | 104 | [160] | |
| TCDCA↓ | \ | \ | \ | 104 | [160] | ||
| PtC↓ | \ | \ | \ | 29 | [14] | ||
| glycine-conjugated bile acids↑ | \ | \ | \ | 29 | [14] | ||
| taurine-conjugated bile acids↑ | \ | \ | \ | 29 | [14] | ||
| PtC↓ | \ | \ | \ | 25 | [161] | ||
| PtC model 1↓ | 80.0% | 95.0% | \ | 25 | [157] | ||
| PtC model2↓ | 88.9% | 87.1% | 87.8% | 49 | [158] | ||
| ON-PC↑ | 85.7% | 80.3% | 86.0% | 46 | [15] | ||
| ON-PC + S-PC | 100.0% | 83.3% | 91.0% | 46 | [15] | CD | |
| EVs source | 5-miR panel | 67.0% | 96.0% | \ | 6 | [170] | |
| miR-10a-5p↑ | \ | \ | \ | 2 | [168] | ||
| miR-181a-5p↑ | \ | \ | \ | 2 | [168] | ||
| circRNA1↑ | \ | \ | 85.7% | 84 | [171] | ||
| ENST00000588480.1↑ | 62.9% | 73.2% | 68.0% | 91 | [173] | ||
| ENST00000517758.1 + ENST00000588480.1 | 82.9% | \ | 70.9% | 91 | [173] | CD | |
| CLDN3↑ | 87.5% | 87.5% | 94.5% | 20 | [175] | ||
| VOS source | VOC model | 90.5% | 72.7% | 89.0% | 32 | [16] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, F.; Liu, J.; Chen, H.; Miao, L.; Xu, Z.; Zhang, G. Diagnosis Biomarkers of Cholangiocarcinoma in Human Bile: An Evidence-Based Study. Cancers 2022, 14, 3921. https://doi.org/10.3390/cancers14163921
Bao F, Liu J, Chen H, Miao L, Xu Z, Zhang G. Diagnosis Biomarkers of Cholangiocarcinoma in Human Bile: An Evidence-Based Study. Cancers. 2022; 14(16):3921. https://doi.org/10.3390/cancers14163921
Chicago/Turabian StyleBao, Fang, Jiayue Liu, Haiyang Chen, Lu Miao, Zhaochao Xu, and Guixin Zhang. 2022. "Diagnosis Biomarkers of Cholangiocarcinoma in Human Bile: An Evidence-Based Study" Cancers 14, no. 16: 3921. https://doi.org/10.3390/cancers14163921
APA StyleBao, F., Liu, J., Chen, H., Miao, L., Xu, Z., & Zhang, G. (2022). Diagnosis Biomarkers of Cholangiocarcinoma in Human Bile: An Evidence-Based Study. Cancers, 14(16), 3921. https://doi.org/10.3390/cancers14163921