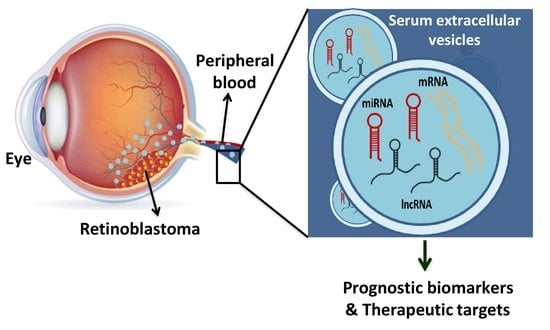
Comprehensive Analysis of Serum Small Extracellular Vesicles-Derived Coding and Non-Coding RNAs from Retinoblastoma Patients for Identifying Regulatory Interactions
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Small Extracellular Vesicles Isolation from Serum
2.3. Transmission Electron Microscopy
2.4. Nanoparticle Tracking Analysis by Zeta View
2.5. Immunoblotting for Exosome Specific Proteins
2.6. RNA Isolation, Library Preparation, Sequencing, and Data Processing
2.7. Differential Expression Analysis
2.8. miRNA and lncRNA Target Analysis
2.9. Functional Enrichment Analysis
2.10. Construction of RNA Interaction Networks
2.11. Construction of Protein-Protein Interaction Network
2.12. Quantitative Reverse Transcriptase-Polymerase Chain Reaction
2.13. Statistical Analysis
3. Results
3.1. Characterization of Serum-Derived Extracellular Vesicles
3.2. Analysis of Serum sEVs RNA Content by RNA Sequencing
3.3. Identification of Differentially Expressed mRNAs, miRNAs and lncRNAs in RB sEVs
3.4. Functional Enrichment Analysis of Differentially Expressed mRNAs and Protein-Protein Interaction-Network of Eye-Related Genes in RB Serum-Derived Small EVs
3.5. Differentially Expressed miRNA-Target Gene Analysis and Functional Enrichment
3.6. miRNA-mRNA Regulatory Network Results
3.7. DE lncRNA Analysis and Functional Enrichment Analysis of Target Genes
| Cell Cycle Specific Genes Dysregulated in RB Serum Small EVs | Fold Change | FDR | No. of Interacting miRNAs (Up Regulated) in RB Serum Small EVs | No. of Interacting miRNAs (Down Regulated) in RB Serum Small EVs | No. of Interacting lncRNAs (Up/Down/N (Neutral) in RB Serum Small EVs |
|---|---|---|---|---|---|
| RB1 | −6.6 | 0.007 | 4 (17-5p, 20a-5p, 132-3p, 215-5p) | 6 (23b-3p, 106b-5p, 192-5p, 130b-3p, 221-3p, 20b-5p) | HOTAIR (N), AATBC (N), MEG3 (N), RB1-DT (N), and PANTR1 (N) |
| CCND1 | 4.03 | 0.005 | 14 (20a-5p, 16-5p, 19a-3p, 17-5p, 425-5P, 155-5p, 24-3p, let-7f-5p, let-7c-5p, let-7a-5p, 98-5p, 101-3p, 342-5p) | 10 (15a-5p, 15b-5p, 106b-5p, 142-5p, 340-5p, 20b-5p, 7706, 323b-3p, let-7e-5p, 7a-3p) | AFAP1-AS1 (Up), DBH-AS1 (Up), MALAT1 (Down) |
| E2F3 | 2.03 | 0.01 | 7 (17-5p, 20a-5p, 101-3p, 24-3p, 16-5p, 660-5p, 425-5P) | 16 (210-3p, 128-3p, 106b-5p, 203a-3p, 221-3p, 32-5p, 30c-5p, 15a-5p, 15b-5p, 92b-3p, 103b, 20b-5p, 4732-3p, 423-5p, 199a-5p, 125a-5p) | FLVCR1-DT (N), NORAD (N) and NEAT1 (N) |
| CDKN1A | −2.2 | 1.0 | 14 (182-5p, 20a-5p, 17-5p, 132-3p, 146b-5p, 10b-5p, 98-5p, let-7f-5p, 7a-5p, 16-5p, 7c-5p, 101-3p, 133a-3p, 181a-5p) | 11 (654-3p, 363-3p, 345-5p, 28-5p, 20b-5p, 125a-5p, 106b-5p, 15a-5p, 15b-5p, 148b-3p, let-7e-5p) | HOTAIR (N), BANCR (Up), DBH-AS1 (Up), HOSA-AS2 MALAT1 (Down), SNHG1 (Down), HOTTIP (Down) MIR31H1G (Down) |
| CDKN1B | −7.6 | 7.5× 10−5 | 181a-5p, 24-3p, 155-5p, 182-5p, | 148-5p | DBH-AS1 (Up) and MALAT1 (Down) |
| TP53 | −1.9 | 1.0 | 11 (16-5p, 10b-5p, 324-5p, 150-5p, 30e-5p, 19b-3p, 20a-5p, 17-5p, 19a-3p, 24-3p, 330-3p) | 10 (125a-5p, 25-3p, 15a-5p, 221-3p, 30c-5p, 106b-5p, 185-5p, 3529-3p, 151a-5p, 28-5p) | MALAT1 (Down) MEG3 (N) SFTA1P (Down), and SNHG1(Down) |
| c-MYC | 1.54 | 1.0 | 14 (24-3p, 98-5p, 155-5p, 17-5p, 20a-5p, 378a-3p, 487b-3p, 19a-3p, 16-5p, 148a-5p, 29a-3p, let-7a-5p, 7c-5p, 7f-5p) | 18 (320b, 744-5p, 423-5p, 323a-3p, 16-2-3p, 7-5p, 126-5p, 25-3p, 106b-5p, 92b-3p, 30c-5p, 23a-3p, 196b-5p, 151a-5p, 125a-5p, let-7e-5p, 130a-3p, 185-5p) | AFAP1-AS1 (Up), PVT1 (N), MALAT1 (Down), RBM5-AS1 (Down), PCATC (N) |
| MYCN | 6.8 | 0.003 | 4 (101-3p, 29a-3p, 19b-3p, 19a-3p) | 4 (let-7e-5p, 126-5p, 144-3p, 103a-3p) | - |
| MDM2 | −6.6 | 0.007 | 6 (17-5P, 20a-5p, 330-3p, 29a-3p, 381-3p, 425-5p | 13 (32-5p, 25-3p, 143-3p, 221-3p, 92b-3p, 363-3p, 20b-5p, 106b-5p, 185-5p, 59, let-7a-3p, 339-5p, 340-5p | MEG3 (N) |
| WASL | −6.9 | 0.002 | 8 (17-5p, 98-5p, let-7f-5p, let-7c-5p, let-7a-5p, 20a-5p, 19a-3p, 19b-3p) | 15 (27b-3p, 379-5p, 148b-3p, 128-3p, 323a-3p, let-7e-5p, 363-3p, 92b-3p, 32-5p, 25-3p, 130b-3p, 130a-3p, 20b-5p, 106b-5p, -590-3p) | SNHG14 (N) and CDKN2B-AS1 (N) |
| HSP90AA11 | −7.4 | 0.0002 | 7 (16-5p, 425-5p, 421, 378a-3p, 30e-5p, 17-5p, 101-3p) | 7 (760, 185-5p, 30c-5p, 25-3p, 889-3p, 148b-3p, 23a-3p) | - |
| XIAP | −7.6 | 0.0001 | 13 (181a-5p, 181b-5p, 215-5p, 101-3p, 17-5p, 24-3p, 20a-5p, 421, 122-5p, 150-5p, 10b-5p, 19b-3p, 19a-3p) | 13 (192-5p, 7-5p, 106b-5p, 20b-5p, 130a-3p, 584-5p, let-7e-5p, 889-3p, 143-3p, 15b-3p, 451b, 130b-3p, 23a-3p) | AFAP1-AS1 (Up), DANCR (N), GHET1 (Down), MALAT1 (Down), PCAT6 (N), PCGEM1 (N), PVT1 (N), and RBM5-AS1 (Down) |
| AKAP8 | −8.1 | 6.1 × 10−6 | 5 (146b-5p, let-7f-5p, let-7c-5p, let-7a-5p, 98-5p) | 2 (92b-3p, let-7e-5p) | - |
| BRCA1 | −2.2 | 0.006 | 5 (16-5p, 24-3p, 215-5p, 181a-5p, 10b-5p) | 3 (15a-5p, 192-5p, 20b-5p) | - |
| CYLD | −2.3 | 0.009 | 5 (17-5p, 16-5p, 20a-5p, 181b-5p, 182-5p) | 6 (106b-5p, 20b-5p, 15b-5p, 15a-5p, 130b-3p, 126-5p) | - |
| FBXO31 | −6.9 | 0.002 | 4 (17-5p, 20a-5p, 3074-5p, 10b-5p) | 7 (192-5p, 92b-3p, 106b-5p, 339-5p, 451b, 3529-3p, 20b-5p) | - |
| KIF2C | 2.5 | 0.001 | 6 (101-3p, 16-5p, 20a-5p, 181a-5p, 181b-5p, 181c-5p) | 2 (148b-3p, 142-5p) | - |
| CEP55 | 7.1 | 0.005 | 5 (155-5p, 215-5p, 16-5p, 19a-3p, 19b-3p) | 10 (192-5p, 103a-3p, 130a-3p, 130b-3p, 148b-3p, 15a-5p, 15b-5p, 411-5p, 199b-3p, 199a-3p) | - |
3.8. LncRNA-miRNA-mRNA Network Results
3.9. Quantitative Reverse Transcriptase-Polymerase Chain Reaction Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Knudson, A.G., Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef]
- Rushlow, D.E.; Mol, B.M.; Kennett, J.Y.; Yee, S.; Pajovic, S.; Thériault, B.L.; Prigoda-Lee, N.L.; Spencer, C.; Dimaras, H.; Corson, T.W.; et al. Characterisation of retinoblastomas without RB1 mutations: Genomic, gene expression, and clinical studies. Lancet Oncol. 2013, 14, 327–334. [Google Scholar] [CrossRef]
- Kaliki, S.; Patel, A.; Iram, S.; Palkonda, V.A.R.; Mohamed, A.; Ramappa, G. Retinoblastoma in India: Clinical presentation and outcome in 1457 patients (2074 eyes). Retina 2019, 39, 379–391. [Google Scholar]
- Berry, J.L.; Xu, L.; Murphree, A.L.; Krishnan, S.; Stachelek, K.; Zolfaghari, E.; McGovern, K.; Kathleen, M.; Carlsson, A.; Kuhn, P.; et al. Potential of Aqueous Humor as a Surrogate Tumor Biopsy for Retinoblastoma. JAMA Ophthalmol. 2017, 135, 1221–1230. [Google Scholar] [CrossRef]
- Tucci, M.; Passarelli, A.; Mannavola, F.; Stucci, L.S.; Ascierto, P.A.; Capone, M.; Madonna, G.; Lopalco, P.; Silvestris, F. Serum exosomes as predictors of clinical response to ipilimumab in metastatic melanoma. OncoImmunology 2018, 7, e1387706. [Google Scholar] [CrossRef]
- Li, S.; Yi, M.; Dong, B.; Tan, X.; Luo, S.; Wu, K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int. J. Cancer 2021, 148, 2640–2651. [Google Scholar] [CrossRef]
- Whiteside, T.L. The potential of tumor-derived exosomes for noninvasive cancer monitoring. Expert Rev. Mol. Diagn. 2015, 15, 1293–1310. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the In-ternational Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef]
- Enderle, D.; Spiel, A.; Coticchia, C.M.; Berghoff, E.; Mueller, R.; Schlumpberger, M.; Sprenger-Haussels, M.; Shaffer, J.M.; Lader, E.; Skog, J.; et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS ONE 2015, 10, e0136133. [Google Scholar]
- Tai, Y.-L.; Chen, K.-C.; Hsieh, J.-T.; Shen, T.-L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.; Enderle, D.; Noerholm, M.; Breakefield, X.; Skog, J. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef]
- Kretschmer, A.; Tutrone, R.; Alter, J.; Berg, E.; Fischer, C.; Kumar, S.; Torkler, P.; Tadigotla, V.; Donovan, M.; Sant, G.; et al. Pre-diagnosis urine exosomal RNA (ExoDx EPI score) is associated with post-prostatectomy pathology outcome. World J. Urol. 2022, 40, 983–989. [Google Scholar] [CrossRef]
- Yu, L. Omics Sequencing of Exosomes in Body Fluids of Patients with Acute Lung Injury; Nanfang Hospital of Southern Medical University: Guangzhou, China, 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT05058768 (accessed on 25 November 2021).
- Cowan, A. Exosome Testing as a Screening Modality for Human Papillomavirus-Positive Oropharyngeal Squamous Cell Carcinoma; University of New Mexico Cancer Center: Albuquerque, NM, USA, 2014. Available online: https://clinicaltrials.gov/ct2/show/NCT02147418 (accessed on 25 November 2021).
- Li, L. Non-Coding RNA in the Exosome of the Epithelia Ovarian Cancer; Peking Union Medical College Hospital: Beijing, China, 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT03738319 (accessed on 25 November 2021).
- Chen, S.; Chen, X.; Qiu, J.; Chen, P.; Han, X.; Wu, Y.; Zhuang, J.; Yang, M.; Wu, C.; Wu, N.; et al. Exosomes derived from retinoblastoma cells enhance tumour deterioration by infiltrating the microenvironment. Oncol. Rep. 2021, 45, 278–290. [Google Scholar] [CrossRef]
- Galardi, A.; Colletti, M.; Lavarello, C.; Di Paolo, V.; Mascio, P.; Russo, I.; Cozza, R.; Romanzo, A.; Valente, P.; De Vito, R.; et al. Proteomic Profiling of Retinoblastoma-Derived Exosomes Reveals Potential Biomarkers of Vitreous Seeding. Cancers 2020, 12, 1555. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Luo, Q.; Liu, X.; Wang, X.; Cui, Z.; He, A.; He, S.; Jiang, Z.; Wu, N.; et al. Retinoblastoma cell-derived exosomes promote angiogenesis of human vesicle endothelial cells through mi-croRNA-92a-3p. Cell Death Dis. 2021, 12, 695. [Google Scholar] [CrossRef]
- Plousiou, M.; De Vita, A.; Miserocchi, G.; Bandini, E.; Vannini, I.; Melloni, M.; Masalu, N.; Fabbri, F.; Serra, P. Growth Inhibition of Retinoblastoma Cell Line by Exosome-Mediated Transfer of miR-142-3p. Cancer Manag. Res. 2022, 14, 2119–2131. [Google Scholar] [CrossRef]
- Ravishankar, H.; Mangani, A.S.; Moses, G.L.P.; Mani, S.P.; Parameswaran, S.; Khetan, V.; Ganesan, S.; Krishnakumar, S. Serum exosomal miRNA as biomarkers for Retinoblastoma. Exp. Eye Res. 2020, 199, 108184. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki. Ethical principles for medical research involving human subjects. Bull. World Health Organ. 2001, 79, 373. [Google Scholar]
- Shields, C.L.; Mashayekhi, A.; Au, A.K.; Czyz, C.; Leahey, A.; Meadows, A.T.; Shields, J.A. The International Classification of Retinoblastoma Predicts Chemoreduction Success. Ophthalmology 2006, 113, 2276–2280. [Google Scholar] [CrossRef]
- Ahmed, F.; Tamma, M.; Pathigadapa, U.; Reddanna, P.; Yenuganti, V.R. Drug Loading and Functional Efficacy of Cow, Buffalo, and Goat Milk-Derived Exosomes: A Comparative Study. Mol. Pharm. 2022, 19, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Mehdiani, A.; Maier, A.; Pinto, A.; Barth, M.; Akhyari, P.; Lichtenberg, A. An Innovative Method for Exosome Quantification and Size Measurement. J. Vis. Exp. 2015, 95, e50974. [Google Scholar] [CrossRef] [PubMed]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Robinson, M.D.; Smyth, G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 2008, 9, 321–332. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J. Educ. Behav. Stat. 2000, 25, 60–83. [Google Scholar] [CrossRef]
- Hsu, S.-D.; Lin, F.-M.; Wu, W.-Y.; Liang, C.; Huang, W.-C.; Chan, W.-L.; Tsai, W.-T.; Chen, G.-Z.; Lee, C.-J.; Chiu, C.-M.; et al. miRTarBase: A database curates experimentally validated microRNA–target interactions. Nucleic Acids Res. 2011, 39, D163–D169. [Google Scholar] [CrossRef]
- Licursi, V.; Conte, F.; Fiscon, G.; Paci, P. MIENTURNET: An interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinform. 2019, 20, 545. [Google Scholar]
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019, 47, D140–D144. [Google Scholar] [CrossRef]
- Zhao, H.; Shi, J.; Zhang, Y.; Xie, A.; Yu, L.; Zhang, C.; Lei, J.; Xu, H.; Leng, Z.; Li, T.; et al. LncTarD: A manually-curated database of experimentally-supported functional lncRNA–target regulations in human diseases. Nucleic Acids Res. 2020, 48, D118–D126. [Google Scholar] [CrossRef] [PubMed]
- RNAcentral Consortium. RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021, 49, D212–D220. [Google Scholar]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Pomaznoy, M.; Ha, B.; Peters, B. GOnet: A tool for interactive Gene Ontology analysis. BMC Bioinform. 2018, 19, 470. [Google Scholar] [CrossRef]
- Kern, F.; Aparicio-Puerta, E.; Li, Y.; Fehlmann, T.; Kehl, T.; Wagner, V.; Ray, K.; Ludwig, N.; Lenhof, H.P.; Meese, E.; et al. miRTargetLink 2.0—interactive miRNA target gene and target pathway networks. Nucleic Acids Res. 2021, 49, W409–W416. [Google Scholar]
- Gerstner, N.; Kehl, T.; Lenhof, K.; Müller, A.; Mayer, C.; Eckhart, L.; Grammes, N.L.; Diener, C.; Hart, M.; Hahn, O.; et al. GeneTrail 3: Advanced high-throughput enrichment analysis. Nucleic Acids Res. 2020, 48, W515–W520. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef] [Green Version]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for Visualization and Analysis of Biological Networks. Comput. Aided Tissue Eng. 2011, 696, 291–303. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [PubMed]
- MacQueen, J. Classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics, Berkeley, CA, USA, 21 June–18 July 1965. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar]
- Lande, K.; Gupta, J.; Ranjan, R.; Kiran, M.; Solis, L.F.T.; Herrera, A.S.; Aliev, G.; Karnati, R. Exosomes: Insights from Retinoblastoma and Other Eye Cancers. Int. J. Mol. Sci. 2020, 21, 7055. [Google Scholar] [CrossRef]
- Deng, F.; Miller, J. A review on protein markers of exosome from different bio-resources and the antibodies used for char-acterization. J. Histotechnol. 2019, 42, 226–239. [Google Scholar]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2, 19671. [Google Scholar] [CrossRef]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the Rna Content of the Exosomes Derived from Blood Serum and Urine and Its Potential as Biomarkers. Philos. Trans. R Soc. Lond. Ser. Biol. Sci. 2014, 369, 20130502. [Google Scholar] [CrossRef]
- König, L.; Kasimir-Bauer, S.; Bittner, A.-K.; Hoffmann, O.; Wagner, B.; Manvailer, L.F.S.; Kimmig, R.; Horn, P.A.; Rebmann, V. Elevated levels of extracellular vesicles are associated with therapy failure and disease progression in breast cancer patients undergoing neoadjuvant chemotherapy. OncoImmunology 2018, 7, e1376153. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLOS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- Corson, T.; Gallie, B. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosom. Cancer 2007, 46, 617–634. [Google Scholar] [CrossRef] [PubMed]
- Vemuganti, G.K.; Balla, M.M.S.; Nair, R.M.; Khan, I.; Kalathur, R.K.R.; Honavar, S.G.; Mohammed, J.A.; Kondaiah, P. Gene expression analysis of retinoblastoma tissues with clinico-histopathologic correlation. J. Radiat. Cancer Res. 2019, 10, 85. [Google Scholar] [CrossRef]
- Rajasekaran, S.; Selvan, L.D.N.; Dotts, K.; Kumar, R.; Rishi, P.; Khetan, V.; Bisht, M.; Sivaraman, K.; Krishnakumar, S.; Sahoo, D.; et al. Non-coding and Coding Transcriptional Profiles Are Significantly Altered in Pediatric Retinoblastoma Tumors. Front. Oncol. 2019, 9, 221. [Google Scholar] [CrossRef]
- Zhao, J.-J.; Yang, J.; Lin, J.; Yao, N.; Zhu, Y.; Zheng, J.; Xu, J.; Cheng, J.Q.; Lin, J.-Y.; Ma, X. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Child’s Nerv. Syst. 2009, 25, 13–20. [Google Scholar] [CrossRef]
- Conkrite, K.; Sundby, M.; Mukai, S.; Thomson, J.M.; Mu, D.; Hammond, S.M.; MacPherson, D. miR-17∼92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011, 25, 1734–1745. [Google Scholar] [CrossRef]
- Huang, J.C.; Babak, T.; Corson, T.; Chua, G.; Khan, S.; Gallie, B.; Hughes, T.R.; Blencowe, B.J.; Frey, B.J.; Morris, Q. Using expression profiling data to identify human microRNA targets. Nat. Chem. Biol. 2007, 4, 1045–1049. [Google Scholar] [CrossRef]
- Beta, M.; Venkatesan, N.; Vasudevan, M.; Vetrivel, U.; Khetan, V.; Krishnakumar, S. Identification and insilico analysis of retinoblastoma serum microRNA profile and gene targets towards pre-diction of novel serum biomarkers. Bioinform. Biol. Insights 2013, 7, BBI.S10501. [Google Scholar]
- Hao, F.; Mou, Y.; Zhang, L.; Wang, S.; Yang, Y. LncRNA AFAP1-AS1 is a prognostic biomarker and serves as oncogenic role in retinoblastoma. Biosci. Rep. 2018, 38, BSR20180384. [Google Scholar] [CrossRef]
- Su, S.; Gao, J.; Wang, T.; Wang, J.; Li, H.; Wang, Z. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in reti-noblastoma. Tumor Biol. 2015, 36, 7205–7211. [Google Scholar]
- Shang, W.; Yang, Y.; Zhang, J.; Wu, Q. Long noncoding RNA BDNF-AS is a potential biomarker and regulates cancer development in human ret-inoblastoma. Biochem. Biophys. Res. Commun. 2018, 497, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Fu, Y.; Lu, X.; Wang, M.; Dong, H.; Li, Q. LncRNA HOTAIR/miR-613/c-met axis modulated epithelial-mesenchymal transition of retinoblastoma cells. J. Cell. Mol. Med. 2018, 22, 5083–5096. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Y.; Xin, X. Long non-coding RNA MALAT1 aggravates human retinoblastoma by sponging miR-20b-5p to upregulate STAT3. Pathol. Res. Pract. 2020, 216, 152977. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Huang, P.; Zhang, J. Hypermethylation of MEG3 promoter correlates with inactivation of MEG3 and poor prognosis in patients with retinoblastoma. J. Transl. Med. 2017, 15, 268. [Google Scholar] [CrossRef]
- Zhong, W.; Yang, J.; Li, M.; Li, L.; Li, A. Long noncoding RNA NEAT1 promotes the growth of human retinoblastoma cells via regulation of miR-204/CXCR4 axis. J. Cell. Physiol. 2019, 234, 11567–11576. [Google Scholar] [CrossRef]
- Cheng, Y.; Chang, Q.; Zheng, B.; Xu, J.; Li, H.; Wang, R. LncRNA XIST promotes the epithelial to mesenchymal transition of retinoblastoma via sponging miR-101. Eur. J. Pharmacol. 2019, 843, 210–216. [Google Scholar] [CrossRef]
- Feng, W.; Zhu, R.; Ma, J.; Song, H. LncRNA ELFN1-AS1 promotes retinoblastoma growth and invasion via regulating miR-4270/SBK1 axis. Cancer Manag. Res. 2021, 13, 1067. [Google Scholar] [CrossRef]
- Sheng, L.; Wu, J.; Gong, X.; Dong, D.; Sun, X. SP1-induced upregulation of lncRNA PANDAR predicts adverse phenotypes in retinoblastoma and regulates cell growth and apoptosis in vitro and in vivo. Gene 2018, 668, 140–145. [Google Scholar] [CrossRef]
- Wang, N.; Fan, H.; Fu, S.; Li, S.; Zhou, B.; Jin, Q.; You, Z. Long noncoding RNA UCA1 promotes carboplatin resistance in retinoblastoma cells by acting as a ceRNA of miR-206. Am. J. Cancer Res. 2022, 12, 2160. [Google Scholar]
- Wang, J.X.; Yang, Y.; Li, K. Long noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9. J. Cell. Physiol. 2018, 233, 6986–6995. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, W.; Li, D.; Li, X.; Huang, J.; Huang, R.; Luo, J. lncRNA FEZF1-AS1 promotes migration, invasion and epithelial-mesenchymal transition of retinoblastoma cells by targeting miR-1236–3p. Mol. Med. Rep. 2020, 22, 3635–3644. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, L.; Lu, L.; Wang, Y. Long Noncoding RNA SNHG16 Sponges miR-182-5p and miR-128-3p To Promote Retinoblastoma Cell Migration and Invasion by Targeting LASP1. OncoTargets Ther. 2019, 12, 8653–8662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, C.; Liu, S.; Lv, Y.; Zhang, C.; Gao, H.; Tan, L.; Wang, H. Long non-coding RNA HOTAIR regulates proliferation and invasion via activating Notch signalling pathway in retinoblastoma. J. Biosci. 2016, 41, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Calin, S.; Yang, Y.; You, M.J.; Calin, G.A. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol. Ther. 2012, 136, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Tang, F.; Li, J.; Yu, H.; Wu, M.; Wu, Y.; Zeng, H.; Hou, K.; Zhang, Q. Tumor-derived exosomes: The emerging orchestrators in melanoma. Biomed. Pharmacother. 2022, 149, 112832. [Google Scholar] [CrossRef]
- Cahill, K.B.; Cote, R.H. Phosphodiesterase 6C, cGMP-specific cone alpha’. AFCS Nat. Mol. Pages 2011, 2011, A001756. [Google Scholar] [CrossRef]
- Ramirez, S.H.; Andrews, A.M.; Paul, D.; Pachter, J.S. Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 2018, 15, 19. [Google Scholar] [CrossRef]
- Li, A.; Zhu, X.; Brown, B.; Craft, C.M. Gene expression networks underlying retinoic acid-induced differentiation of human retinoblastoma cells. Investig. Opthalmology Vis. Sci. 2003, 44, 996–1007. [Google Scholar] [CrossRef]
- Gómez-Romero, L.; Alvarez-Suarez, D.E.; Hernández-Lemus, E.; Ponce-Castañeda, M.V.; Tovar, H. The regulatory landscape of retinoblastoma: A pathway analysis perspective. R Soc. Open Sci. 2022, 9, 220031. [Google Scholar] [CrossRef]
- Aldiri, I.; Xu, B.; Wang, L.; Chen, X.; Hiler, D.; Griffiths, L.; Valentine, M.; Shirinifard, A.; Thiagarajan, S.; Sablauer, A.; et al. The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron 2017, 94, 550–568.e10. [Google Scholar] [CrossRef]
- Lee, C.; Kim, J.K. Chromatin regulators in retinoblastoma: Biological roles and therapeutic applications. J. Cell. Physiol. 2021, 236, 2318–2332. [Google Scholar] [CrossRef] [PubMed]
- Chivukula, R.R.; Mendell, J.T. Circular reasoning: MicroRNAs and cell-cycle control. Trends Biochem. Sci. 2008, 33, 474–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, R.; Zou, H.; Wang, L.F.; Song, M.J.; Liu, L.; Zhang, H. Identification of microRNA-mRNA regulatory networks and pathways related to retinoblastoma across human and mouse. Int. J. Ophthalmol. 2020, 13, 535. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Kitagawa, K.; Kotake, Y.; Niida, H.; Ohhata, T. Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 2013, 70, 4785–4794. [Google Scholar] [CrossRef]
- Shang, Y. LncRNA THOR acts as a retinoblastoma promoter through enhancing the combination of c-myc mRNA and IGF2BP1 protein. Biomed. Pharmacother. 2018, 106, 1243–1249. [Google Scholar] [CrossRef]
- Ni, W.; Li, Z.; Ai, K. lncRNA ZFPM2-AS1 promotes retinoblastoma progression by targeting microRNA miR-511-3p/paired box protein 6 (PAX6) axis. Bioengineered 2022, 13, 1637–1649. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, X.-W. The silencing of long non-coding RNA ANRIL suppresses invasion, and promotes apoptosis of ret-inoblastoma cells through the ATM-E2F1 signaling pathway. Biosci. Rep. 2018, 38, BSR20180558. [Google Scholar] [CrossRef]
- Chen, X.; Wang, C.-C.; Guan, N.-N. Computational Models in Non-Coding RNA and Human Disease. Int. J. Mol. Sci. 2020, 21, 1557. [Google Scholar] [CrossRef]
- Ren, H.; Guo, X.; Li, F.; Xia, Q.; Chen, Z.; Xing, Y. Four Autophagy-Related Long Noncoding RNAs Provide Coexpression and ceRNA Mechanisms in Reti-noblastoma through Bioinformatics and Experimental Evidence. ACS Omega 2021, 6, 33976–33984. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, X.; Zhang, M.; Shen, Q.; Qin, Z. Identification of hub genes and pathways associated with retinoblastoma based on co-expression network analysis. Genet. Mol. Res. 2015, 14, 16151–16161. [Google Scholar] [CrossRef]
- Ganguly, A.; Shields, C.L. Differential gene expression profile of retinoblastoma compared to normal retina. Mol. Vis. 2010, 16, 1292–1303. [Google Scholar] [PubMed]







| S No | Gene | Function | Log2 (Fold Change) | FDR |
|---|---|---|---|---|
| 1 | PAX4 (Paired Box 4) | Retina development in camera type eye | 4.5 | 3.1× 10−7 |
| 2 | WNT5A (Wnt Family Member 5A) | Optic cup formation involved in camera type eye development | 8.5 | 3.9× 10−7 |
| 3 | INHBA (Inhibin Subunit Beta A) | Eyelid development in camera type eye | 3.4 | 0.0001 |
| 4 | PFDN5 (Prefoldin Subunit 5) | Retina development in camera type eye | 7.4 | 0.0003 |
| 5 | RARB (Retinoic Acid Receptor Beta) | Embryonic eye morphogenesis | 2.8 | 0.0004 |
| 6 | RBP4 (Retinol Binding Protein 4) | Eye development | 7.3 | 0.0005 |
| 7 | ALDH1A2 (Aldehyde Dehydrogenase 1 Family Member A2) | Embryonic camera type eye development | 2.6 | 0.0006 |
| 8 | TWSG1 (Twisted Gastrulation BMP Signaling Modulator 1) | Camera type eye development | 4.42 | 0.001 |
| 9 | BHLHE23 (Basic Helix-Loop-Helix Family Member E23) | Post embryonic eye morphogenesis | 7.1 | 0.001 |
| 10 | MEIS3 (Meis Homeobox 3) | Eye development | 3.1 | 0.001 |
| 11 | TULP1 (TUB Like Protein 1) | Retina development in camera type eye | 2.5 | 0.001 |
| 12 | OLFM3 (Olfactomedin 3) | Eye photoreceptor cell development | 2.7 | 0.002 |
| 13 | CYP1B1 (Cytochrome P450 Family 1 Subfamily B Member 1) | Retina vasculature development in camera type eye | 6.9 | 0.002 |
| 14 | PDE6B (Phosphodiesterase 6B) | Retina development in camera type eye | 3.0 | 0.002 |
| 15 | PTN (Pleiotrophin) | Retina development in camera type eye | 2.4 | 0.01 |
| 16 | PROX1 (Prospero Homeobox 1) | Retina morphogenesis in camera type eye | 2.7 | 0.01 |
| 17 | CYP1A1 (Cytochrome P450 Family 1 Subfamily A Member 1) | Camera type eye development | 2.9 | 0.01 |
| 18 | ACHE (Acetylcholinesterase) | Retina development in camera type eye | 2.1 | 0.01 |
| 19 | PDGFRA (Platelet Derived Growth Factor Receptor Alpha) | Retina vasculature development in camera type eye | 2.2 | 0.04 |
| 20 | BMPR1B (Bone Morphogenetic Protein Receptor Type 1B) | Retina development in camera type eye | 2.5 | 0.05 |
| S No | Gene | Biological Function | Log2 (Fold Change) | FDR |
|---|---|---|---|---|
| 1 | SOX8 (Sex Determining Region Y) Transcription Factor 8) | Negative regulation of photoreceptor cell differentiation, Retina development in camera type eye | −7.4 | 0.0003 |
| 2 | SPATA7 (Spermatogenesis Associated 7) | Photoreceptor cell maintenance | −7.2 | 0.0007 |
| 3 | OPN3 (Opsin 3) | Phototransduction | −5.9 | 0.04 |
| 4 | PDE6C (Phosphodiesterase 6C) | Phototransduction visible light | −6.7 | 0.005 |
| 5 | RP1 (Retinitis Pigmentosa 1 Axonemal Microtubule Associated) | phototransduction, visible light, Retina development in camera type eye | −3.9 | 0.0001 |
| 6 | IFT20 (Intraflagellar Transport 20) | Photoreceptor cell outer segment organization | −7.2 | 0.0005 |
| 7 | BAK1 (BCL2 Antagonist/Killer 1) | Post embryonic camera type eye morphogenesis | −6.5 | 0.01 |
| 8 | CTNS (Cystinosin, Lysosomal Cystine Transporter) | Lens development in camera-type eye | −7.0 | 0.001 |
| 9 | PAX2 (Paired Box 2) | Optic cup morphogenesis involved in camera type eye development | −4.6 | 0.0001 |
| 10 | GNB1 (G Protein Subunit Beta 1) | Retina development in camera-type eye | −7.8 | 3.1× 10−5 |
| 11 | CRYBG3 (Crystallin Beta-Gamma Domain Containing 3) | Lens development in camera type eye | −7.5 | 0.0001 |
| 12 | XRN2 (5′-3′ Exoribonuclease 2) | Retina development in camera type eye | −6.8 | 0.003 |
| 13 | YY1 (Transcription Factor) | Camera type eye morphogenesis | −7.8 | 4.3× 10−5 |
| 14 | BMP7 (Bone Morphogenetic Protein 7) | Embryonic camera type eye morphogenesis | −2.8 | 0.005 |
| 15 | HSF4 (Heat Shock Transcription Factor 4) | Camera type eye development | −7.2 | 0.0007 |
| 16 | CALB1 (Calbindin 1) | Retina development in camera type eye | −6.8 | 0.003 |
| 17 | PBX4 (PBX Homeobox 4) | Eye development | −5.9 | 0.04 |
| 18 | SLC1A1 (Solute Carrier Family 1 Member 1) | Retina development in camera type eye | −6.5 | 0.01 |
| 19 | GATA3 (GATA Binding Protein 3) | Lens development in camera type eye | −7.1 | 0.001 |
| Gene | Closeness Centrality | Betweenness Centrality | Degree Layout |
|---|---|---|---|
| MALAT1 | 0.45 | 0.5 | 42 |
| HOTAIR | 0.4 | 0.22 | 25 |
| NEAT1 | 0.35 | 0.2 | 24 |
| AFAP1-AS1 | 0.36 | 0.2 | 23 |
| MEG3 | 0.35 | 0.1 | 15 |
| SNHG1 | 0.34 | 0.1 | 13 |
| CDKN1A | 0.36 | 0.08 | 8 |
| MIR145 | 0.39 | 0.08 | 6 |
| EZH2 | 0.37 | 0.04 | 5 |
| ZEB1 | 0.39 | 0.06 | 5 MIR101 0.31 0.02 4 BCL2 0.33 0.05 6 TP53 0.36 0.04 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manukonda, R.; Yenuganti, V.R.; Nagar, N.; Dholaniya, P.S.; Malpotra, S.; Attem, J.; Reddy, M.M.; Jakati, S.; Mishra, D.K.; Reddanna, P.; et al. Comprehensive Analysis of Serum Small Extracellular Vesicles-Derived Coding and Non-Coding RNAs from Retinoblastoma Patients for Identifying Regulatory Interactions. Cancers 2022, 14, 4179. https://doi.org/10.3390/cancers14174179
Manukonda R, Yenuganti VR, Nagar N, Dholaniya PS, Malpotra S, Attem J, Reddy MM, Jakati S, Mishra DK, Reddanna P, et al. Comprehensive Analysis of Serum Small Extracellular Vesicles-Derived Coding and Non-Coding RNAs from Retinoblastoma Patients for Identifying Regulatory Interactions. Cancers. 2022; 14(17):4179. https://doi.org/10.3390/cancers14174179
Chicago/Turabian StyleManukonda, Radhika, Vengala Rao Yenuganti, Nupur Nagar, Pankaj Singh Dholaniya, Shivani Malpotra, Jyothi Attem, Mamatha M. Reddy, Saumya Jakati, Dilip K Mishra, Pallu Reddanna, and et al. 2022. "Comprehensive Analysis of Serum Small Extracellular Vesicles-Derived Coding and Non-Coding RNAs from Retinoblastoma Patients for Identifying Regulatory Interactions" Cancers 14, no. 17: 4179. https://doi.org/10.3390/cancers14174179
APA StyleManukonda, R., Yenuganti, V. R., Nagar, N., Dholaniya, P. S., Malpotra, S., Attem, J., Reddy, M. M., Jakati, S., Mishra, D. K., Reddanna, P., Poluri, K. M., Vemuganti, G. K., & Kaliki, S. (2022). Comprehensive Analysis of Serum Small Extracellular Vesicles-Derived Coding and Non-Coding RNAs from Retinoblastoma Patients for Identifying Regulatory Interactions. Cancers, 14(17), 4179. https://doi.org/10.3390/cancers14174179