Tumor Colonization and Therapy by Escherichia coli Nissle 1917 Strain in Syngeneic Tumor-Bearing Mice Is Strongly Affected by the Gut Microbiome
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterium and Mammalian Cell Cultures
2.2. PCR and Western Blot Analyses of the Bacterial Strain EcN/pMUT-gfp Knr
2.2.1. PCR Analysis
2.2.2. Western Blot Analysis
2.3. Antibiotic-Induced Microbiome Depletion in Mice
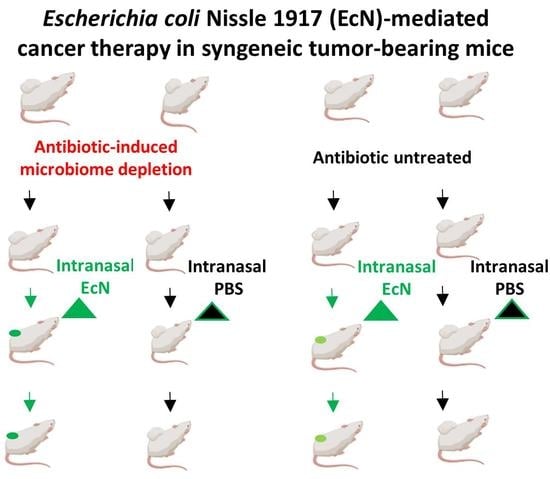
2.4. Animal Experiments
2.5. Analysis of Bacterial Colony Forming Units (CFU) in Feces and Tumor Samples
2.6. Multiplexed Immunohistochemistry (CODEX) of Tumor Tissue Sections
2.7. Statistical Analysis
3. Results
3.1. Effects of Antibiotic-Induced Gut Microbiome Depletion and Intranasal (i.n.) EcN/pMUT-gfp Knr Applications on Tumor Growth in 4T1 /BALB/c Syngeneic Mouse Model
3.2. Distribution of EcN/pMUT-gfp Knr in Syngenic 4T1 Tumor Bearing BALB/c Mice
3.2.1. Distribution of EcN/pMUT-gfp Knr Strain in Tumors
3.2.2. Presence of EcN/pMUT-gfp Knr in Fecal Samples of Mice with Intranasal Bacterium Injections
3.3. Role of Host Immune System in the Antitumor Mechanism of EcN/pMUT-gfp Knr Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bermudes, D.; Low, K.B.; Pawelek, J.; Feng, M.; Belcourt, M.; Zheng, L.-M.; King, I. Tumour-SelectiveSalmonella-BasedCancer Therapy. Biotechnol. Genet. Eng. Rev. 2001, 18, 219–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, T.; Fujimori, M.; Hamaji, Y.; Hama, Y.; Ito, K.-I.; Amano, J.; Taniguchi, S. Genetically engineered Bifidobacterium longum for tumor-targeting enzyme-prodrug therapy of autochthonous mammary tumors in rats. Cancer Sci. 2006, 97, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.A.; Shabahang, S.; Timiryasova, T.M.; Zhang, Q.; Beltz, R.; Gentschev, I.; Goebel, W.; Szalay, A.A. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat. Biotechnol. 2004, 22, 313–320. [Google Scholar] [CrossRef]
- Wood, L.M.; Guirnalda, P.D.; Seavey, M.M.; Paterson, Y. Cancer immunotherapy using Listeria monocytogenes and listerial virulence factors. Immunol. Res. 2008, 42, 233–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, N.; Bettegowda, C.; Cheong, I.; Geschwind, J.-F.; Drake, C.G.; Hipkiss, E.L.; Tatsumi, M.; Dang, L.H.; Diaz, L.A.; Pomper, M.; et al. Bacteriolytic therapy can generate a potent immune response against experimental tumors. Proc. Natl. Acad. Sci. USA 2004, 101, 15172–15177. [Google Scholar] [CrossRef] [Green Version]
- Theys, J.; Pennington, O.; Dubois, L.; Anlezark, G.; Vaughan, T.; Mengesha, A.; Landuyt, W.; Anné, J.; Burke, P.J.; Dûrre, P.; et al. Repeated cycles of Clostridium-directed enzyme prodrug therapy result in sustained antitumour effects in vivo. Br. J. Cancer 2006, 95, 1212–1219. [Google Scholar] [CrossRef] [Green Version]
- Duong, M.T.-Q.; Qin, Y.; You, S.-H.; Min, J.-J. Bacteria-cancer interactions: Bacteria-based cancer therapy. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Pawelek, J.M.; Low, K.B.; Bermudes, D. Bacteria as tumour-targeting vectors. Lancet Oncol. 2003, 4, 548–556. [Google Scholar] [CrossRef]
- Zhou, S.; Gravekamp, C.; Bermudes, D.; Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 2018, 18, 727–743. [Google Scholar] [CrossRef]
- Schmidt, C.K.; Medina-Sánchez, M.; Edmondson, R.J.; Schmidt, O.G. Engineering microrobots for targeted cancer therapies from a medical perspective. Nat. Commun. 2020, 11, 5618. [Google Scholar] [CrossRef]
- Basu, P.; Mehta, A.; Jain, M.; Gupta, S.; Nagarkar, R.V.; John, S.; Petit, R. A Randomized Phase 2 Study of ADXS11-001 Listeria monocytogenes–Listeriolysin O Immunotherapy With or Without Cisplatin in Treatment of Advanced Cervical Cancer. Int. J. Gynecol. Cancer 2018, 28, 764–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://clinicaltrials.gov/ct2/show/record/NCT02853604 (accessed on 20 September 2022).
- Blum-Oehler, G.; Oswald, S.; Eiteljörge, K.; Sonnenborn, U.; Schulze, J.; Kruis, W.; Hacker, J. Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples. Res. Microbiol. 2003, 154, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Rembacken, B.J.; Snelling, A.M.; Hawkey, P.M.; Chalmers, D.M.; Axon, A. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: A randomised trial. Lancet 1999, 354, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Chmielewska, A.; Szajewska, H. Systematic review of randomised controlled trials: Probiotics for functional constipation. World J. Gastroenterol. 2010, 16, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Malchow, H.A. Crohn’s Disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn’s disease? J. Clin. Gastroenterol. 1997, 25, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, M.R.; Fuschi, D.; Cremon, C.; Carapelle, M.; Dino, P.; Marcellini, M.M.; Dothel, G.; De Ponti, F.; Stanghellini, V.; Barbara, G. Escherichia coli Nissle 1917 restores epithelial permeability alterations induced by irritable bowel syndrome mediators. Neurogastroenterol. Motil. 2018, 30, e13388. [Google Scholar] [CrossRef]
- Isabella, V.M.; Ha, B.N.; Castillo, M.J.; Lubkowicz, D.J.; Rowe, S.E.; Millet, Y.A.; Anderson, C.L.; Li, N.; Fisher, A.B.; West, K.A.; et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018, 36, 857–864. [Google Scholar] [CrossRef]
- Ou, B.; Yang, Y.; Tham, W.L.; Chen, L.; Guo, J.; Zhu, G. Genetic engineering of probiotic Escherichia coli Nissle 1917 for clinical application. Appl. Microbiol. Biotechnol. 2016, 100, 8693–8699. [Google Scholar] [CrossRef]
- Available online: https://clinicaltrials.gov/ct2/show/NCT04167137 (accessed on 25 October 2022).
- Gronbach, K.; Eberle, U.; Muller, M.; Olschlager, T.A.; Dobrindt, U.; Leithauser, F.; Niess, J.H.; Doring, G.; Reimann, J.; Autenrieth, I.B.; et al. Safety of Probiotic Escherichia coli Strain Nissle 1917 Depends on Intestinal Microbiota and Adaptive Immunity of the Host. Infect. Immun. 2010, 78, 3036–3046. [Google Scholar] [CrossRef] [Green Version]
- Stritzker, J.; Weibel, S.; Hill, P.J.; Oelschlaeger, T.A.; Goebel, W.; Szalay, A.A. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int. J. Med. Microbiol. 2007, 297, 151–162. [Google Scholar] [CrossRef]
- Stritzker, J.; Weibel, S.; Seubert, C.; Götz, A.; Tresch, A.; van Rooijen, N.; Oelschlaeger, T.A.; Hill, P.; Gentschev, I.; Szalay, A.A. Enterobacterial tumor colonization in mice depends on bacterial metabolism and macrophages but is independent of chemotaxis and motility. Int. J. Med. Microbiol. 2010, 300, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Westphal, K.; Leschner, S.; Jablonska, J.; Loessner, H.; Weiss, S. Containment of Tumor-Colonizing Bacteria by Host Neutrophils. Cancer Res. 2008, 68, 2952–2960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, Y.; Xia, L.; Zhang, X.; Ding, X.; Yan, F.; Wu, F. Escherichia coli Nissle 1917 Targets and Restrains Mouse B16 Melanoma and 4T1 Breast Tumors through Expression of Azurin Protein. Appl. Environ. Microbiol. 2012, 78, 7603–7610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leventhal, D.S.; Sokolovska, A.; Li, N.; Plescia, C.; Kolodziej, S.A.; Gallant, C.W.; Christmas, R.; Gao, J.-R.; James, M.J.; Abin-Fuentes, A.; et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 2020, 11, 2739. [Google Scholar] [CrossRef]
- Altenhoefer, A.; Oswald, S.; Sonnenborn, U.; Enders, C.; Schulze, J.; Hacker, J.; Oelschlaeger, T.A. The probiotic Escherichia colistrain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol. Med. Microbiol. 2004, 40, 223–229. [Google Scholar] [CrossRef] [Green Version]
- Dexter, D.L.; Kowalski, H.M.; Blazar, B.A.; Fligiel, Z.; Vogel, R.; Heppner, G.H. Heterogeneity of tumor cells from a single mouse mammary tumor. Cancer Res. 1978, 38, 3174–3181. [Google Scholar]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [Green Version]
- Costello, E.K.; Stagaman, K.; Dethlefsen, L.; Bohannan, B.J.M.; Relman, D.A. The Application of Ecological Theory Toward an Understanding of the Human Microbiome. Science 2012, 336, 1255–1262. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.Y.; Yin, T.L.; Zhou, J.; Xu, J.; Lu, X.J. Gut microbiome and cancer immunotherapy. J. Cell. Physiol. 2020, 235, 4082–4088. [Google Scholar] [CrossRef]
- Cheng, W.Y.; Wu, C.-Y.; Yu, J. The role of gut microbiota in cancer treatment: Friend or foe? Gut 2020, 69, 1867–1876. [Google Scholar] [CrossRef]
- Black, S.; Phillips, D.; Hickey, J.W.; Kennedy-Darling, J.; Venkataraaman, V.G.; Samusik, N.; Goltsev, Y.; Schürch, C.M.; Nolan, G.P. CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat. Protoc. 2021, 16, 3802–3835. [Google Scholar] [CrossRef] [PubMed]
- Goltsev, Y.; Samusik, N.; Kennedy-Darling, J.; Bhate, S.; Hale, M.; Vazquez, G.; Black, S.; Nolan, G.P. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 2018, 174, 968–981.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reikvam, D.H.; Erofeev, A.; Sandvik, A.; Grcic, V.; Jahnsen, F.L.; Gaustad, P.; McCoy, K.D.; MacPherson, A.J.; Meza-Zepeda, L.A.; Johansen, F.-E. Depletion of Murine Intestinal Microbiota: Effects on Gut Mucosa and Epithelial Gene Expression. PLoS ONE 2011, 6, e17996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarrinpar, A.; Chaix, A.; Xu, Z.Z.; Chang, M.W.; Marotz, C.A.; Saghatelian, A.; Knight, R.; Panda, S. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 2018, 9, 2872. [Google Scholar] [CrossRef] [Green Version]
- Seo, E.-J.; Weibel, S.; Wehkamp, J.; Oelschlaeger, T.A. Construction of recombinant E. coli Nissle 1917 (EcN) strains for the expression and secretion of defensins. Int. J. Med. Microbiol. 2012, 302, 276–287. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 6 July 2021).
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef] [Green Version]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Hollister, E.B.; Riehle, K.; Luna, R.A.; Weidler, E.M.; Rubio-Gonzales, M.; Mistretta, T.-A.; Raza, S.; Doddapaneni, H.V.; Metcalf, G.A.; Muzny, D.M.; et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome 2015, 3, 36. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.S.B.; Raes, J.; Bork, P. The Human Gut Microbiome: From Association to Modulation. Cell 2018, 172, 1198–1215. [Google Scholar] [CrossRef] [Green Version]
- Sarate, P.J.; Heinl, S.; Poiret, S.; Drinić, M.; Zwicker, C.; Schabussova, I.; Daniel, C.; Wiedermann, U.E. coli Nissle 1917 is a safe mucosal delivery vector for a birch-grass pollen chimera to prevent allergic poly-sensitization. Mucosal Immunol. 2019, 12, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Schürch, C.M.; Bhate, S.S.; Barlow, G.L.; Phillips, D.J.; Noti, L.; Zlobec, I.; Chu, P.; Black, S.; Demeter, J.; McIlwain, D.R.; et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 2020, 182, 1341–1359.e19. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, J.A.; Huang, C.-J.; Doody, A.M.; Leung, T.; Mineta, K.; Feng, D.D.; Wayne, E.C.; Nishimura, N.; Leifer, C.; DeLisa, M.; et al. Mechanistic Insight into the TH1-Biased Immune Response to Recombinant Subunit Vaccines Delivered by Probiotic Bacteria-Derived Outer Membrane Vesicles. PLoS ONE 2014, 9, e112802. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Coria, L.; Rosemblit, C.; Rivas, M.A.; Proietti, C.J.; Flaqué, M.C.D.; Beguelin, W.; Frahm, I.; Charreau, E.H.; Cassataro, J.; et al. Targeting Stat3 Induces Senescence in Tumor Cells and Elicits Prophylactic and Therapeutic Immune Responses against Breast Cancer Growth Mediated by NK Cells and CD4+T Cells. J. Immunol. 2012, 189, 1162–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weibel, S.; Stritzker, J.; Eck, M.; Goebel, W.; Szalay, A.A. Colonization of experimental murine breast tumours by Escherichia coli K-12 significantly alters the tumour microenvironment. Cell. Microbiol. 2008, 10, 1235–1248. [Google Scholar] [CrossRef]
- Yu, X.; Lin, C.; Yu, J.; Qi, Q.; Wang, Q. Bioengineered Escherichia coliNissle 1917 for tumour-targeting therapy. Microb. Biotechnol. 2020, 13, 629–636. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lee, H.K. The Role of Gut Microbiota in Modulating Tumor Growth and Anticancer Agent Efficacy. Mol. Cells 2021, 44, 356–362. [Google Scholar] [CrossRef]







| Product Code | Product (Target—Dye-Reporter) | Vendor |
|---|---|---|
| 4230019 | Ki67(AKYP0052)-BX047—Atto 550 | Akoya Biosciences, Inc. |
| 4250016 | CD4(AKYP0041)-BX026—Atto 550 | Akoya Biosciences, Inc |
| 4350014 | CD3(17A2)-BX021—Cy5 | Akoya Biosciences, Inc |
| 4250014 | CD19(AKYP0033)-BX020—Atto 550 | Akoya Biosciences, Inc |
| 4250017 | CD8a(AKYP0044)-BX029—Atto 550 | Akoya Biosciences, Inc |
| 4250003 | MHC II(AKYP0006)-BX014—Atto 550 | Akoya Biosciences, Inc |
| 4450015 | CD11b(AKYP0040)-BX025—Alexa Fluor™ 750 | Akoya Biosciences, Inc |
| 4550108 | CD11c(AKYP0045)-BX030—Cy5 | Akoya Biosciences, Inc |
| 4550102 | CD49f(AKYP0018)-BX033—Cy5 | Akoya Biosciences, Inc |
| 4450002 | CD45(AKYP0005)-BX007—Alexa Fluor™ 750 | Akoya Biosciences, Inc |
| Primers | Origin DNA | DNA Sequences | Length |
|---|---|---|---|
| 4L2 4R2 | Chromosomal | 5′-GGG CGA TCG GAT TTA ATC AT-3′ 5′-CGA GGA CTC GGA GCT TAC TG-3′ | 186 bp |
| 5L1 5R1 | Chromosomal | 5′-GCC TCT CGC AAC TTA ACG AC-3′ 5′-AGT TAT CCA GCG TTG CCA TC-3′ | 232 bp |
| Muta5 Muta6 | Plasmid pMUT1 | 5′-AAC TGT GAA GCG ATG AAC CC-3′ 5′-GGA CTG TTC AGA GAG CTA TC-3′ | 361 bp |
| Muta7 Muta8 | Plasmid pMUT2 | 5′-GAC CAA GCG ATA ACC GGA TG-3‘ 5′-GTG AGA TGA TGG CCA CGA TT-3′ | 427 bp |
| Muta9 Muta10 | Plasmid pMUT2 | 5′-GCG AGG TAA CCT CGA ACA TG-3′ 5′-CGG CGT ATC GAT AAT TCA CG-3′ | 313 bp |
| Time Point (Day)/CFU/g Feces | Day 0 before ABT | 4 Days after ABT | 7 Days after ABT |
|---|---|---|---|
| CFUs on LB agar plates | 1.76 × 105 ± 4.82 × 104 | 3.19 × 104 ± 3.05 × 103 | n.d. |
| CFUs on Sabouraud agar plates | 3.27 × 103 ± 8.16 × 102 | n.d. | n.d. |
| CFUs on Schaedler agar plates | 2.62 × 107 ± 3.78 × 106 | 1.60 × 105 ± 1.05 × 105 | n.d. |
| Mouse No. | CFU /per 1g Tumor Tissue |
|---|---|
| Group 1/Antibiotics pretreated (ATB)/EcN | |
| 206/ABT | 9.64 × 102 ± 2.14 × 102 |
| 207/ABT | 1.82 × 102 |
| 208/ABT | 7.37 × 103 ± 3.39 × 102 |
| 209/ABT | 1.60 × 103 |
| 210/ABT | 6.2 × 10 |
| Group 2/No antibiotics/EcN | |
| 291 | 9.2 × 10 |
| 290 | n.d. |
| 289 | n.d. |
| 288 | 1.18 × 102 |
| 287 | n.d. |
| Mice Group/ Day of Treatment | Group 1 ABT */EcN 11dpbi | Group1 ABT*/EcN 18dpbi | Group 2/EcN 11dpbi | Group 2/EcN 18dpbi |
|---|---|---|---|---|
| CFU per100 mg feces | 1.8 × 103 ± 4.0 × 102 | 9.0 × 102 ± 1.0 × 101 | 1.0 × 102 | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gentschev, I.; Petrov, I.; Ye, M.; Kafuri Cifuentes, L.; Toews, R.; Cecil, A.; Oelschaeger, T.A.; Szalay, A.A. Tumor Colonization and Therapy by Escherichia coli Nissle 1917 Strain in Syngeneic Tumor-Bearing Mice Is Strongly Affected by the Gut Microbiome. Cancers 2022, 14, 6033. https://doi.org/10.3390/cancers14246033
Gentschev I, Petrov I, Ye M, Kafuri Cifuentes L, Toews R, Cecil A, Oelschaeger TA, Szalay AA. Tumor Colonization and Therapy by Escherichia coli Nissle 1917 Strain in Syngeneic Tumor-Bearing Mice Is Strongly Affected by the Gut Microbiome. Cancers. 2022; 14(24):6033. https://doi.org/10.3390/cancers14246033
Chicago/Turabian StyleGentschev, Ivaylo, Ivan Petrov, Mingyu Ye, Lina Kafuri Cifuentes, Romy Toews, Alexander Cecil, Tobias A. Oelschaeger, and Aladar A. Szalay. 2022. "Tumor Colonization and Therapy by Escherichia coli Nissle 1917 Strain in Syngeneic Tumor-Bearing Mice Is Strongly Affected by the Gut Microbiome" Cancers 14, no. 24: 6033. https://doi.org/10.3390/cancers14246033
APA StyleGentschev, I., Petrov, I., Ye, M., Kafuri Cifuentes, L., Toews, R., Cecil, A., Oelschaeger, T. A., & Szalay, A. A. (2022). Tumor Colonization and Therapy by Escherichia coli Nissle 1917 Strain in Syngeneic Tumor-Bearing Mice Is Strongly Affected by the Gut Microbiome. Cancers, 14(24), 6033. https://doi.org/10.3390/cancers14246033