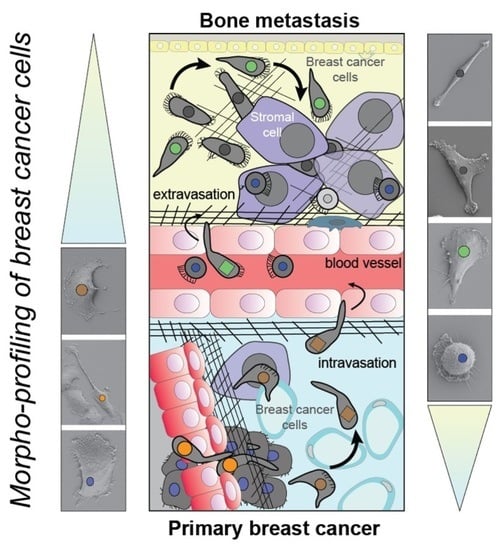
Decoding Single Cell Morphology in Osteotropic Breast Cancer Cells for Dissecting Their Migratory, Molecular and Biophysical Heterogeneity
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Antibodies
2.3. Cell Culture
2.4. Time-Lapse Video Microscopy
2.5. Fluorescence Labeling, Lectin Cytochemistry and Laser Scanning Confocal Microscopy
2.6. Scanning Electron Microscopy
2.7. Atomic Force Microscopy Indentation
2.8. Real-Time Deformability Cytometry
2.9. Flow Cytometry
2.10. Immunoblotting
2.11. Transcriptome
2.12. RNA-Seq Data Analysis
2.13. Statistical Analysis
3. Results
3.1. MB-231 Cells and Their Bone-Seeking Derivatives Exhibit a Broad Spectrum of Single Cell Morphotypes
3.2. Differential Motility of Breast Cancer Cells with Specific Morphotypes
3.3. MET Cells, but Not MB-231 and BONE, Form Colonies That Affect Their Motility
3.4. Differential Effects of CXCL12 and EGF on Cell Motility and Morphotype Distribution
3.5. Differential Expression of Selective Cluster of Differentiation Molecules among Breast Cancer Cell Sub-Lines
3.6. Differential Surface Glycosylation Patterns among Breast Cancer Cell Sub-Lines
3.7. Phenotypic Comparison of Bone vs. Brain Metastatic Breast Cancer Cells
3.8. Differential Biophysical Properties of Single Cell Morphotypes
3.9. Transcriptional Profiling of Breast Cancer Cell Sub-Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic behavior of breast cancer subtypes. J. Clin. Oncol. 2010, 28, 3271–3277. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet 2005, 365, 1687–1717. [Google Scholar] [CrossRef]
- Coleman, R.E.; Rubens, R.D. The clinical course of bone metastases from breast cancer. Br. J. Cancer 1987, 55, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Mundy, G.R.; Yoneda, T. Facilitation and suppression of bone metastasis. Clin. Orthop. Relat. Res. 1995, 312, 34–44. [Google Scholar]
- D’Oronzo, S.; Coleman, R.; Brown, J.; Silvestris, F. Metastatic bone disease: Pathogenesis and therapeutic options: Up-date on bone metastasis management. J. Bone Oncol. 2019, 15, 100205. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast cancer subtypes predict the preferential site of distant metastases: A SEER based study. Oncotarget 2017, 8, 27990–27996. [Google Scholar] [CrossRef] [Green Version]
- Paget, S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989, 8, 98–101. [Google Scholar]
- Zlotnik, A.; Burkhardt, A.M.; Homey, B. Homeostatic chemokine receptors and organ-specific metastasis. Nat. Rev. Immunol. 2011, 11, 597–606. [Google Scholar] [CrossRef]
- Yoneda, T.; Williams, P.J.; Hiraga, T.; Niewolna, M.; Nishimura, R. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J. Bone Miner. Res. 2001, 16, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Cailleau, R.; Olivé, M.; Cruciger, Q.V. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. In Vitro 1978, 14, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.H.; Gilkes, D.M.; Phillip, J.M.; Narkar, A.; Cheng, T.W.; Marchand, J.; Lee, M.H.; Li, R.; Wirtz, D. Single-cell morphology encodes metastatic potential. Sci. Adv. 2020, 6, eaaw6938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmons, J.K.; Hildreth, B.E., 3rd; Supsavhad, W.; Elshafae, S.M.; Hassan, B.B.; Dirksen, W.P.; Toribio, R.E.; Rosol, T.J. Animal Models of Bone Metastasis. Vet. Pathol. 2015, 52, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendre, M.S.; Gaddy-Kurten, D.; Mon-Foote, T.; Akel, N.S.; Skinner, R.A.; Nicholas, R.W.; Suva, L.J. Expression of interleukin 8 and not parathyroid hormone-related protein by human breast cancer cells correlates with bone metastasis in vivo. Cancer Res. 2002, 62, 5571–5579. [Google Scholar] [PubMed]
- Shafiee, A.; McGovern, J.A.; Lahr, C.A.; Meinert, C.; Moi, D.; Wagner, F.; Landgraf, M.; De-Juan-Pardo, E.; Mazzieri, R.; Hutmacher, D.W. Immune system augmentation via humanization using stem/progenitor cells and bioengineering in a breast cancer model study. Int. J. Cancer 2018, 143, 1470–1482. [Google Scholar] [CrossRef] [Green Version]
- Carcel-Trullols, J.; Stanley, J.S.; Saha, R.; Shaaf, S.; Bendre, M.S.; Monzavi-Karbassi, B.; Suva, L.J.; Kieber-Emmons, T. Characterization of the glycosylation profile of the human breast cancer cell line, MDA-231, and a bone colonizing variant. Int. J. Oncol. 2006, 28, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Göbel, A.; Thiele, S.; Browne, A.J.; Rauner, M.; Zinna, V.M.; Hofbauer, L.C.; Rachner, T.D. Combined inhibition of the mevalonate pathway with statins and zoledronic acid potentiates their anti-tumor effects in human breast cancer cells. Cancer Lett. 2016, 375, 162–171. [Google Scholar] [CrossRef]
- Göbel, A.; Browne, A.J.; Thiele, S.; Rauner, M.; Hofbauer, L.C.; Rachner, T.D. Potentiated suppression of Dickkopf-1 in breast cancer by combined administration of the mevalonate pathway inhibitors zoledronic acid and statins. Breast Cancer Res. Treat. 2015, 154, 623–631. [Google Scholar] [CrossRef]
- Bos, P.D.; Zhang, X.H.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tinevez, J.Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Freund, D.; Bauer, N.; Boxberger, S.; Feldmann, S.; Streller, U.; Ehninger, G.; Werner, C.; Bornhäuser, M.; Oswald, J.; Corbeil, D. Polarization of human hematopoietic progenitors during contact with multipotent mesenchymal stromal cells: Effects on proliferation and clonogenicity. Stem Cells Dev. 2006, 15, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Hertz, H. Ueber die Berühung fester elastischer Koerper. J. für die Reine und Angew. Math. 1882, 1882, 156–171. [Google Scholar] [CrossRef]
- Sneddon, I.N. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- Radmacher, M. Studying the mechanics of cellular processes by atomic force microscopy. Methods Cell Biol. 2007, 83, 347–372. [Google Scholar] [CrossRef] [PubMed]
- Otto, O.; Rosendahl, P.; Mietke, A.; Golfier, S.; Herold, C.; Klaue, D.; Girardo, S.; Pagliara, S.; Ekpenyong, A.; Jacobi, A.; et al. Real-time deformability cytometry: On-the-fly cell mechanical phenotyping. Nat. Methods 2015, 12, 199–202. [Google Scholar] [CrossRef]
- Mietke, A.; Otto, O.; Girardo, S.; Rosendahl, P.; Taubenberger, A.; Golfier, S.; Ulbricht, E.; Aland, S.; Guck, J.; Fischer-Friedrich, E. Extracting Cell Stiffness from Real-Time Deformability Cytometry: Theory and Experiment. Biophys. J. 2015, 109, 2023–2036. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Small, J.V.; Anderson, K.; Rottner, K. Actin and the coordination of protrusion, attachment and retraction in cell crawling. Biosci. Rep. 1996, 16, 351–368. [Google Scholar] [CrossRef]
- McNeil, E.; Capaldo, C.T.; Macara, I.G. Zonula occludens-1 function in the assembly of tight junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell 2006, 17, 1922–1932. [Google Scholar] [CrossRef] [Green Version]
- Frixen, U.H.; Behrens, J.; Sachs, M.; Eberle, G.; Voss, B.; Warda, A.; Löchner, D.; Birchmeier, W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 1991, 113, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Guriec, N.; Marcellin, L.; Gairard, B.; Caldéroli, H.; Wilk, A.; Renaud, R.; Bergerat, J.P.; Oberling, F. E-cadherin mRNA expression in breast carcinomas correlates with overall and disease-free survival. Invasion Metastasis 1996, 16, 19–26. [Google Scholar] [PubMed]
- Muscarella, A.M.; Dai, W.; Mitchell, P.G.; Zhang, W.; Wang, H.; Jia, L.; Stossi, F.; Mancini, M.A.; Chiu, W.; Zhang, X.H. Unique cellular protrusions mediate breast cancer cell migration by tethering to osteogenic cells. NPJ Breast Cancer 2020, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Nieman, M.T.; Prudoff, R.S.; Johnson, K.R.; Wheelock, M.J. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J. Cell Biol. 1999, 147, 631–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roger, L.; Jullien, L.; Gire, V.; Roux, P. Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon cancer cells. J. Cell Sci. 2010, 123, 1295–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chekhun, S.; Bezdenezhnykh, N.; Shvets, J.; Lukianova, N. Expression of biomarkers related to cell adhesion, metastasis and invasion of breast cancer cell lines of different molecular subtype. Exp. Oncol. 2013, 35, 174–179. [Google Scholar] [PubMed]
- Sun, X.; Cheng, G.; Hao, M.; Zheng, J.; Zhou, X.; Zhang, J.; Taichman, R.S.; Pienta, K.J.; Wang, J. CXCL12 / CXCR4 / CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010, 29, 709–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, T.; Resat, H. Quantitative investigation of MDA-MB-231 breast cancer cell motility: Dependence on epidermal growth factor concentration and its gradient. Mol. Biosyst. 2017, 13, 2069–2082. [Google Scholar] [CrossRef]
- Lu, Z.; Jiang, G.; Blume-Jensen, P.; Hunter, T. Epidermal growth factor-induced tumor cell invasion and metastasis initiated by dephosphorylation and downregulation of focal adhesion kinase. Mol. Cell. Biol. 2001, 21, 4016–4031. [Google Scholar] [CrossRef] [Green Version]
- Bailly, M.; Macaluso, F.; Cammer, M.; Chan, A.; Segall, J.E.; Condeelis, J.S. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 1999, 145, 331–345. [Google Scholar] [CrossRef] [Green Version]
- Suraneni, P.; Rubinstein, B.; Unruh, J.R.; Durnin, M.; Hanein, D.; Li, R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 2012, 197, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [Green Version]
- Radke, J.; Roßner, F.; Redmer, T. CD271 determines migratory properties of melanoma cells. Sci. Rep. 2017, 7, 9834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Hayashida, T.; Forster, N.; Li, C.; Shen, D.; Maheswaran, S.; Chen, L.; Anderson, K.S.; Ellisen, L.W.; Sgroi, D.; et al. The integrin alpha(v)beta(3-5) ligand MFG-E8 is a p63/p73 target gene in triple-negative breast cancers but exhibits suppressive functions in ER(+) and erbB2(+) breast cancers. Cancer Res. 2011, 71, 937–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Li, J.; Song, Q.; Zhu, K.; Yu, X.; Tian, Y.; Zhang, J. Reduction in milk fat globule-EGF factor 8 inhibits triple-negative breast cancer cell viability and migration. Oncol. Lett. 2019, 17, 3457–3465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Zhou, S.M.; Cheng, L.; Guo, S.J.; Wang, Y.; Czajkowsky, D.M.; Gao, H.; Hu, X.F.; Tao, S.C. Lectin RCA-I specifically binds to metastasis-associated cell surface glycans in triple-negative breast cancer. Breast Cancer Res. 2015, 17, 36. [Google Scholar] [CrossRef] [Green Version]
- Lv, J.; Liu, Y.; Cheng, F.; Li, J.; Zhou, Y.; Zhang, T.; Zhou, N.; Li, C.; Wang, Z.; Ma, L.; et al. Cell softness regulates tumorigenicity and stemness of cancer cells. EMBO J. 2021, 40, e106123. [Google Scholar] [CrossRef]
- Gensbittel, V.; Kräter, M.; Harlepp, S.; Busnelli, I.; Guck, J.; Goetz, J.G. Mechanical Adaptability of Tumor Cells in Metastasis. Dev. Cell 2021, 56, 164–179. [Google Scholar] [CrossRef]
- Sanz-Moreno, V.; Marshall, C.J. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr. Opin. Cell Biol. 2010, 22, 690–696. [Google Scholar] [CrossRef]
- Yin, Z.; Sadok, A.; Sailem, H.; McCarthy, A.; Xia, X.; Li, F.; Garcia, M.A.; Evans, L.; Barr, A.R.; Perrimon, N.; et al. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nat. Cell Biol. 2013, 15, 860–871. [Google Scholar] [CrossRef] [Green Version]
- Friedl, P.; Wolf, K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat. Rev. Cancer 2003, 3, 362–374. [Google Scholar] [CrossRef]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell migration: Integrating signals from front to back. Science 2003, 302, 1704–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.H.; Phillip, J.M.; Khatau, S.B.; Chen, W.C.; Stirman, J.; Rosseel, S.; Tschudi, K.; Van Patten, J.; Wong, M.; Gupta, S.; et al. Evolution of cellular morpho-phenotypes in cancer metastasis. Sci. Rep. 2015, 5, 18437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedl, P.; Wolf, K. Plasticity of cell migration: A multiscale tuning model. J. Cell Biol. 2010, 188, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.; Thiery, J.P. Epithelial-mesenchymal transitions: Insights from development. Development 2012, 139, 3471–3486. [Google Scholar] [CrossRef] [Green Version]
- Bouchet, B.P.; Akhmanova, A. Microtubules in 3D cell motility. J. Cell Sci. 2017, 130, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Schillaci, O.; Fontana, S.; Monteleone, F.; Taverna, S.; Di Bella, M.A.; Di Vizio, D.; Alessandro, R. Exosomes from metastatic cancer cells transfer amoeboid phenotype to non-metastatic cells and increase endothelial permeability: Their emerging role in tumor heterogeneity. Sci. Rep. 2017, 7, 4711. [Google Scholar] [CrossRef] [Green Version]
- Shafqat-Abbasi, H.; Kowalewski, J.M.; Kiss, A.; Gong, X.; Hernandez-Varas, P.; Berge, U.; Jafari-Mamaghani, M.; Lock, J.G.; Strömblad, S. An analysis toolbox to explore mesenchymal migration heterogeneity reveals adaptive switching between distinct modes. eLife 2016, 5, e11384. [Google Scholar] [CrossRef] [PubMed]
- Abercrombie, M.; Heaysman, J.E.; Pegrum, S.M. The locomotion of fibroblasts in culture. I. Movements of the leading edge. Exp. Cell Res. 1970, 59, 393–398. [Google Scholar] [CrossRef]
- Gerisch, G.; Keller, H.U. Chemotactic reorientation of granulocytes stimulated with micropipettes containing fMet-Leu-Phe. J. Cell Sci. 1981, 52, 1–10. [Google Scholar] [CrossRef]
- Parent, C.A.; Blacklock, B.J.; Froehlich, W.M.; Murphy, D.B.; Devreotes, P.N. G protein signaling events are activated at the leading edge of chemotactic cells. Cell 1998, 95, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Insall, R.H. Understanding eukaryotic chemotaxis: A pseudopod-centred view. Nat. Rev. Mol. Cell Biol. 2010, 11, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Mazo, I.; Leung, H.; Engelke, K.; von Andrian, U.H.; Deryugina, E.I.; Strongin, A.Y.; Bröcker, E.B.; Friedl, P. Compensation mechanism in tumor cell migration: Mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003, 160, 267–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keren, K.; Pincus, Z.; Allen, G.M.; Barnhart, E.L.; Marriott, G.; Mogilner, A.; Theriot, J.A. Mechanism of shape determination in motile cells. Nature 2008, 453, 475–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorotnikov, A.V.; Tyurin-Kuzmin, P.A. Chemotactic signaling in mesenchymal cells compared to amoeboid cells. Genes Dis. 2014, 1, 162–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.J.; Le Berre, M.; Lautenschlaeger, F.; Maiuri, P.; Callan-Jones, A.; Heuzé, M.; Takaki, T.; Voituriez, R.; Piel, M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 2015, 160, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Fackler, O.T.; Grosse, R. Cell motility through plasma membrane blebbing. J. Cell Biol. 2008, 181, 879–884. [Google Scholar] [CrossRef] [Green Version]
- Charras, G.; Paluch, E. Blebs lead the way: How to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol. 2008, 9, 730–736. [Google Scholar] [CrossRef]
- Paluch, E.K.; Raz, E. The role and regulation of blebs in cell migration. Curr. Opin. Cell Biol. 2013, 25, 582–590. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, K.; Soldati, T. Dissection of amoeboid movement into two mechanically distinct modes. J. Cell Sci. 2006, 119, 3833–3844. [Google Scholar] [CrossRef] [Green Version]
- Torka, R.; Thuma, F.; Herzog, V.; Kirfel, G. ROCK signaling mediates the adoption of different modes of migration and invasion in human mammary epithelial tumor cells. Exp. Cell Res. 2006, 312, 3857–3871. [Google Scholar] [CrossRef] [PubMed]
- Lämmermann, T.; Sixt, M. Mechanical modes of ‘amoeboid’ cell migration. Curr. Opin. Cell Biol. 2009, 21, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, A.V.; Freund, D.; Bornhäuser, M.; Corbeil, D. Polarization and migration of hematopoietic stem and progenitor cells rely on the RhoA/ROCK I pathway and an active reorganization of the microtubule network. J. Biol. Chem. 2010, 285, 31661–31671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, P.Y.; Huttenlocher, A. Interstitial leukocyte migration in vivo. Curr. Opin. Cell Biol. 2013, 25, 650–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyun, Y.M.; Sumagin, R.; Sarangi, P.P.; Lomakina, E.; Overstreet, M.G.; Baker, C.M.; Fowell, D.J.; Waugh, R.E.; Sarelius, I.H.; Kim, M. Uropod elongation is a common final step in leukocyte extravasation through inflamed vessels. J. Exp. Med. 2012, 209, 1349–1362. [Google Scholar] [CrossRef] [Green Version]
- Butcher, E.C.; Picker, L.J. Lymphocyte homing and homeostasis. Science 1996, 272, 60–66. [Google Scholar] [CrossRef]
- Luster, A.D.; Alon, R.; von Andrian, U.H. Immune cell migration in inflammation: Present and future therapeutic targets. Nat. Immunol. 2005, 6, 1182–1190. [Google Scholar] [CrossRef]
- Heasman, S.J.; Carlin, L.M.; Cox, S.; Ng, T.; Ridley, A.J. Coordinated RhoA signaling at the leading edge and uropod is required for T cell transendothelial migration. J. Cell Biol. 2010, 190, 553–563. [Google Scholar] [CrossRef] [Green Version]
- Poincloux, R.; Collin, O.; Lizárraga, F.; Romao, M.; Debray, M.; Piel, M.; Chavrier, P. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc. Natl. Acad. Sci. USA 2011, 108, 1943–1948. [Google Scholar] [CrossRef] [Green Version]
- Guzman, A.; Avard, R.C.; Devanny, A.J.; Kweon, O.S.; Kaufman, L.J. Delineating the role of membrane blebs in a hybrid mode of cancer cell invasion in three-dimensional environments. J. Cell Sci. 2020, 133, jcs236778. [Google Scholar] [CrossRef]
- Francis, K.; Ramakrishna, R.; Holloway, W.; Palsson, B.O. Two new pseudopod morphologies displayed by the human hematopoietic KG1a progenitor cell line and by primary human CD34(+) cells. Blood 1998, 92, 3616–3623. [Google Scholar] [CrossRef] [PubMed]
- Kuçi, S.; Wessels, J.T.; Bühring, H.J.; Schilbach, K.; Schumm, M.; Seitz, G.; Löffler, J.; Bader, P.; Schlegel, P.G.; Niethammer, D.; et al. Identification of a novel class of human adherent CD34- stem cells that give rise to SCID-repopulating cells. Blood 2003, 101, 869–876. [Google Scholar] [CrossRef]
- Münsterberg, J.; Loreth, D.; Brylka, L.; Werner, S.; Karbanová, J.; Gandrass, M.; Schneegans, S.; Besler, K.; Hamester, F.; Robador, J.R.; et al. ALCAM contributes to brain metastasis formation in non-small-cell lung cancer through interaction with the vascular endothelium. Neuro Oncol. 2020, 22, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Rappa, G.; Green, T.M.; Karbanová, J.; Corbeil, D.; Lorico, A. Tetraspanin CD9 determines invasiveness and tumorigenicity of human breast cancer cells. Oncotarget 2015, 6, 7970–7991. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Takahashi, K. WAVE2- and microtubule-dependent formation of long protrusions and invasion of cancer cells cultured on three-dimensional extracellular matrices. Cancer Sci. 2008, 99, 2252–2259. [Google Scholar] [CrossRef]
- Schoumacher, M.; Goldman, R.D.; Louvard, D.; Vignjevic, D.M. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 2010, 189, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Mader, C.C.; Oser, M.; Magalhaes, M.A.; Bravo-Cordero, J.J.; Condeelis, J.; Koleske, A.J.; Gil-Henn, H. An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 2011, 71, 1730–1741. [Google Scholar] [CrossRef] [Green Version]
- Xue, C.; Wyckoff, J.; Liang, F.; Sidani, M.; Violini, S.; Tsai, K.L.; Zhang, Z.Y.; Sahai, E.; Condeelis, J.; Segall, J.E. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006, 66, 192–197. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Lorenz, M.; Kempiak, S.; Sarmiento, C.; Coniglio, S.; Symons, M.; Segall, J.; Eddy, R.; Miki, H.; Takenawa, T.; et al. Molecular mechanisms of invadopodium formation: The role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 2005, 168, 441–452. [Google Scholar] [CrossRef] [Green Version]
- Sutoh Yoneyama, M.; Hatakeyama, S.; Habuchi, T.; Inoue, T.; Nakamura, T.; Funyu, T.; Wiche, G.; Ohyama, C.; Tsuboi, S. Vimentin intermediate filament and plectin provide a scaffold for invadopodia, facilitating cancer cell invasion and extravasation for metastasis. Eur. J. Cell Biol. 2014, 93, 157–169. [Google Scholar] [CrossRef]
- Whipple, R.A.; Balzer, E.M.; Cho, E.H.; Matrone, M.A.; Yoon, J.R.; Martin, S.S. Vimentin filaments support extension of tubulin-based microtentacles in detached breast tumor cells. Cancer Res. 2008, 68, 5678–5688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraley, S.I.; Feng, Y.; Krishnamurthy, R.; Kim, D.H.; Celedon, A.; Longmore, G.D.; Wirtz, D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 2010, 12, 598–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyanagi, J.; Ogawa, T.; Sato, H.; Higashi, S.; Miyazaki, K. Epithelial-mesenchymal transition stimulates human cancer cells to extend microtubule-based invasive protrusions and suppresses cell growth in collagen gel. PLoS ONE 2012, 7, e53209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eddy, R.J.; Weidmann, M.D.; Sharma, V.P.; Condeelis, J.S. Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis. Trends Cell Biol. 2017, 27, 595–607. [Google Scholar] [CrossRef]
- Weaver, A.M. Invadopodia: Specialized cell structures for cancer invasion. Clin. Exp. Metastasis 2006, 23, 97–105. [Google Scholar] [CrossRef]
- Linder, S.; Wiesner, C.; Himmel, M. Degrading devices: Invadosomes in proteolytic cell invasion. Annu. Rev. Cell Dev. Biol. 2011, 27, 185–211. [Google Scholar] [CrossRef]
- Bauer, N.; Fonseca, A.V.; Florek, M.; Freund, D.; Jászai, J.; Bornhäuser, M.; Fargeas, C.A.; Corbeil, D. New insights into the cell biology of hematopoietic progenitors by studying prominin-1 (CD133). Cells Tissues Organs 2008, 188, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Sanz-Moreno, V.; Marshall, C.J. The metastasis gene NEDD9 product acts through integrin beta3 and Src to promote mesenchymal motility and inhibit amoeboid motility. J. Cell Sci. 2012, 125, 1814–1826. [Google Scholar] [CrossRef] [Green Version]
- Jones, B.C.; Kelley, L.C.; Loskutov, Y.V.; Marinak, K.M.; Kozyreva, V.K.; Smolkin, M.B.; Pugacheva, E.N. Dual Targeting of Mesenchymal and Amoeboid Motility Hinders Metastatic Behavior. Mol. Cancer Res. 2017, 15, 670–682. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Moreno, V.; Gadea, G.; Ahn, J.; Paterson, H.; Marra, P.; Pinner, S.; Sahai, E.; Marshall, C.J. Rac activation and inactivation control plasticity of tumor cell movement. Cell 2008, 135, 510–523. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, M.S.; Parsons, J.T.; Bouton, A.H. Cas and NEDD9 Contribute to Tumor Progression through Dynamic Regulation of the Cytoskeleton. Genes Cancer 2012, 3, 371–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, C.; Wang, C.; Wang, L.; Ma, M.; Niu, C.; Sun, X.; Du, J.; Dong, Z.; Zhu, S.; Lu, J.; et al. NEDD9 is a positive regulator of epithelial-mesenchymal transition and promotes invasion in aggressive breast cancer. PLoS ONE 2011, 6, e22666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLaughlin, S.L.; Ice, R.J.; Rajulapati, A.; Kozyulina, P.Y.; Livengood, R.H.; Kozyreva, V.K.; Loskutov, Y.V.; Culp, M.V.; Weed, S.A.; Ivanov, A.V.; et al. NEDD9 depletion leads to MMP14 inactivation by TIMP2 and prevents invasion and metastasis. Mol. Cancer Res. 2014, 12, 69–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.M.; Deng, X.; Gong, W.; Su, S. Chemokines and their role in tumor growth and metastasis. J. Immunol. Methods 1998, 220, 1–17. [Google Scholar] [CrossRef]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Wendel, C.; Hemping-Bovenkerk, A.; Krasnyanska, J.; Mees, S.T.; Kochetkova, M.; Stoeppeler, S.; Haier, J. CXCR4/CXCL12 participate in extravasation of metastasizing breast cancer cells within the liver in a rat model. PLoS ONE 2012, 7, e30046. [Google Scholar] [CrossRef] [Green Version]
- Peled, A.; Petit, I.; Kollet, O.; Magid, M.; Ponomaryov, T.; Byk, T.; Nagler, A.; Ben-Hur, H.; Many, A.; Shultz, L.; et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science 1999, 283, 845–848. [Google Scholar] [CrossRef] [Green Version]
- López-Gil, J.C.; Martin-Hijano, L.; Hermann, P.C.; Sainz, B., Jr. The CXCL12 Crossroads in Cancer Stem Cells and Their Niche. Cancers 2021, 13, 469. [Google Scholar] [CrossRef]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordón-Cardo, C.; Guise, T.A.; Massagué, J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003, 3, 537–549. [Google Scholar] [CrossRef] [Green Version]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef] [Green Version]
- Shields, J.D.; Fleury, M.E.; Yong, C.; Tomei, A.A.; Randolph, G.J.; Swartz, M.A. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell 2007, 11, 526–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, M.R.; Hou, M.F.; Chang, H.C.; Hung, W.C. Cyclooxygenase-2 up-regulates CCR7 via EP2/EP4 receptor signaling pathways to enhance lymphatic invasion of breast cancer cells. J. Biol. Chem. 2008, 283, 11155–11163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legler, D.F.; Uetz-von Allmen, E.; Hauser, M.A. CCR7: Roles in cancer cell dissemination, migration and metastasis formation. Int. J. Biochem. Cell Biol. 2014, 54, 78–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellahcène, A.; Bachelier, R.; Detry, C.; Lidereau, R.; Clézardin, P.; Castronovo, V. Transcriptome analysis reveals an osteoblast-like phenotype for human osteotropic breast cancer cells. Breast Cancer Res. Treat. 2007, 101, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Patsialou, A.; Wang, Y.; Lin, J.; Whitney, K.; Goswami, S.; Kenny, P.A.; Condeelis, J.S. Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012, 14, R139. [Google Scholar] [CrossRef] [Green Version]
- Karagiannis, G.S.; Goswami, S.; Jones, J.G.; Oktay, M.H.; Condeelis, J.S. Signatures of breast cancer metastasis at a glance. J. Cell Sci. 2016, 129, 1751–1758. [Google Scholar] [CrossRef] [Green Version]
- Baskaran, J.P.; Weldy, A.; Guarin, J.; Munoz, G.; Shpilker, P.H.; Kotlik, M.; Subbiah, N.; Wishart, A.; Peng, Y.; Miller, M.A.; et al. Cell shape, and not 2D migration, predicts extracellular matrix-driven 3D cell invasion in breast cancer. APL Bioeng. 2020, 4, 026105. [Google Scholar] [CrossRef]
- Meyer, A.S.; Hughes-Alford, S.K.; Kay, J.E.; Castillo, A.; Wells, A.; Gertler, F.B.; Lauffenburger, D.A. 2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen. J. Cell Biol. 2012, 197, 721–729. [Google Scholar] [CrossRef] [Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bemmerlein, L.; Deniz, I.A.; Karbanová, J.; Jacobi, A.; Drukewitz, S.; Link, T.; Göbel, A.; Sevenich, L.; Taubenberger, A.V.; Wimberger, P.; et al. Decoding Single Cell Morphology in Osteotropic Breast Cancer Cells for Dissecting Their Migratory, Molecular and Biophysical Heterogeneity. Cancers 2022, 14, 603. https://doi.org/10.3390/cancers14030603
Bemmerlein L, Deniz IA, Karbanová J, Jacobi A, Drukewitz S, Link T, Göbel A, Sevenich L, Taubenberger AV, Wimberger P, et al. Decoding Single Cell Morphology in Osteotropic Breast Cancer Cells for Dissecting Their Migratory, Molecular and Biophysical Heterogeneity. Cancers. 2022; 14(3):603. https://doi.org/10.3390/cancers14030603
Chicago/Turabian StyleBemmerlein, Lila, Ilker A. Deniz, Jana Karbanová, Angela Jacobi, Stephan Drukewitz, Theresa Link, Andy Göbel, Lisa Sevenich, Anna V. Taubenberger, Pauline Wimberger, and et al. 2022. "Decoding Single Cell Morphology in Osteotropic Breast Cancer Cells for Dissecting Their Migratory, Molecular and Biophysical Heterogeneity" Cancers 14, no. 3: 603. https://doi.org/10.3390/cancers14030603
APA StyleBemmerlein, L., Deniz, I. A., Karbanová, J., Jacobi, A., Drukewitz, S., Link, T., Göbel, A., Sevenich, L., Taubenberger, A. V., Wimberger, P., Kuhlmann, J. D., & Corbeil, D. (2022). Decoding Single Cell Morphology in Osteotropic Breast Cancer Cells for Dissecting Their Migratory, Molecular and Biophysical Heterogeneity. Cancers, 14(3), 603. https://doi.org/10.3390/cancers14030603