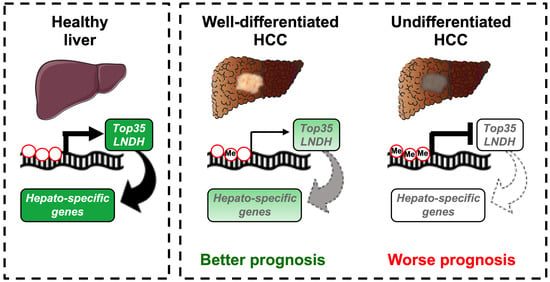
DNA Methylation Regulates a Set of Long Non-Coding RNAs Compromising Hepatic Identity during Hepatocarcinogenesis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Samples
2.2. Public Datasets
2.3. Cell Lines Culture and Transfection
2.4. Total DNA Isolation
2.5. Targeted Bisulfite Sequencing
2.6. Total RNA Isolation
2.7. RT-PCR and qPCR
2.8. Prediction of lncRNA Function
2.9. Statistical Analysis
3. Results
3.1. Identification of a Set of lncRNAs Downregulated in HCC
3.2. Downregulation of the Top35 LNDH through Promoter DNA Methylation
3.3. The Top35 LNDH Are Preferentially Expressed in Adult Liver Tissue
3.4. A Subset of the Top35 LNDH Play a Role in Liver Differentiation
3.5. The Expression of the Top35 LNDH Correlates with Tumor Grading and Patients’ Overall Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berasain, C.; Avila, M.A. Regulation of Hepatocyte Identity and Quiescence. Cell Mol. Life Sci. 2015, 72, 3831–3851. [Google Scholar] [CrossRef] [PubMed]
- Martins-Filho, S.N.; Paiva, C.; Azevedo, R.S.; Alves, V.A.F. Histological Grading of Hepatocellular Carcinoma—A Systematic Review of Literature. Front. Med. 2017, 4, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, K.; Matsuda, M.; Ohkura, Y.; Kawamura, Y.; Inoue, M.; Hashimoto, M.; Ikeda, K.; Kumada, H.; Watanabe, G. The Influence of Histological Differentiation Grade on the Outcome of Liver Resection for Hepatocellular Carcinomas 2 cm or Smaller in Size. World J. Surg. 2015, 39, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, G.P.; Pinelli, D.; Benedetto, F.D.; Marini, E.; Corno, V.; Guizzetti, M.; Aluffi, A.; Zambelli, M.; Fagiuoli, S.; Lucà, M.G.; et al. Predictive Value of Nodule Size and Differentiation in HCC Recurrence after Liver Transplantation. Surg. Oncol. 2016, 25, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Wright, A.E.; Mank, J.E. The Evolution of Gene Expression and the Transcriptome–Phenotype Relationship. Semin. Cell Dev. Biol. 2012, 23, 222–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, G.; Heard, E. Advances in Epigenetics Link Genetics to the Environment and Disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brien, G.L.; Valerio, D.G.; Armstrong, S.A. Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs. Cancer Cell 2016, 29, 464–476. [Google Scholar] [CrossRef] [Green Version]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic Plasticity and the Hallmarks of Cancer. Science 2017, 357, eaal2380. [Google Scholar] [CrossRef] [Green Version]
- Meunier, L.; Hirsch, T.Z.; Caruso, S.; Imbeaud, S.; Bayard, Q.; Roehrig, A.; Couchy, G.; Nault, J.; Llovet, J.M.; Blanc, J.; et al. DNA Methylation Signatures Reveal the Diversity of Processes Remodeling Hepatocellular Carcinoma Methylomes. Hepatology 2021, 74, 816–834. [Google Scholar] [CrossRef]
- Mallardo, M.; Poltronieri, P.; D’Urso, O.F. Non-Protein Coding RNA Biomarkers and Differential Expression in Cancers: A Review. J. Exp. Clin. Cancer Res. Cr. 2008, 27, 19. [Google Scholar] [CrossRef] [Green Version]
- Unfried, J.P.; Serrano, G.; Suárez, B.; Sangro, P.; Ferretti, V.; Prior, C.; Boix, L.; Bruix, J.; Sangro, B.; Segura, V.; et al. Identification of Coding and Long Noncoding RNAs Differentially Expressed in Tumors and Preferentially Expressed in Healthy Tissues. Cancer Res. 2019, 79, 5167–5180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Repáraz, D.; Ruiz, M.; Llopiz, D.; Silva, L.; Vercher, E.; Aparicio, B.; Egea, J.; Tamayo-Uria, I.; Hervás-Stubbs, S.; García-Balduz, J.; et al. Neoantigens as Potential Vaccines in Hepatocellular Carcinoma. J. Immunother. Cancer 2022, 10, e003978. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Sandoval, J.; Heyn, H.; Moran, S.; Serra-Musach, J.; Pujana, M.A.; Bibikova, M.; Esteller, M. Validation of a DNA Methylation Microarray for 450,000 CpG Sites in the Human Genome. Epigenetics 2011, 6, 692–702. [Google Scholar] [CrossRef]
- Laurent, V.; Glaise, D.; Nübel, T.; Gilot, D.; Corlu, A.; Loyer, P. Highly Efficient SiRNA and Gene Transfer into Hepatocyte-like HepaRG Cells and Primary Human Hepatocytes: New Means for Drug Metabolism and Toxicity Studies. Methods Mol. Biol. 2013, 987, 295–314. [Google Scholar] [CrossRef]
- Arechederra, M.; Daian, F.; Yim, A.; Bazai, S.K.; Richelme, S.; Dono, R.; Saurin, A.J.; Habermann, B.H.; Maina, F. Hypermethylation of Gene Body CpG Islands Predicts High Dosage of Functional Oncogenes in Liver Cancer. Nat. Commun. 2018, 9, 3164. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, T.-Y.; Anderton, L.C.; Cao, Q.; Yang, R. LncGSEA: A Versatile Tool to Infer LncRNA Associated Pathways from Large-Scale Cancer Transcriptome Sequencing Data. BMC Genom. 2021, 22, 574. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, L.; Gu, J.; Zhang, H.; Yuan, J.; Lian, Q.; Lv, G.; Wang, S.; Wu, Y.; Yang, Y.-C.T.; et al. Recurrently Deregulated LncRNAs in Hepatocellular Carcinoma. Nat. Commun. 2017, 8, 14421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteller, M. Epigenetic Changes in Cancer. F1000 Biol. Rep. 2011, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Zhi, H.; Li, X.; Wang, P.; Gao, Y.; Gao, B.; Zhou, D.; Zhang, Y.; Guo, M.; Yue, M.; Shen, W.; et al. Lnc2Meth: A Manually Curated Database of Regulatory Relationships between Long Non-Coding RNAs and DNA Methylation Associated with Human Disease. Nucleic Acids Res. 2018, 46, D133–D138. [Google Scholar] [CrossRef] [PubMed]
- Gailhouste, L.; Liew, L.C.; Yasukawa, K.; Hatada, I.; Tanaka, Y.; Nakagama, H.; Ochiya, T. Differentiation Therapy by Epigenetic Reconditioning Exerts Antitumor Effects on Liver Cancer Cells. Mol. Ther. 2018, 26, 1840–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gailhouste, L.; Liew, L.C.; Yasukawa, K.; Hagiwara, K.; Iwazaki, N.; Yamada, Y.; Hatada, I.; Ochiya, T. Epigenetic Reprogramming of Human Hepatoma Cells: A Low-Cost Option for Drug Metabolism Assessment. Cell Mol. Gastroenterol. Hepatol. 2017, 5, 454–457. [Google Scholar] [CrossRef] [Green Version]
- Dannenberg, L.O.; Edenberg, H.J. Epigenetics of Gene Expression in Human Hepatoma Cells: Expression Profiling the Response to Inhibition of DNA Methylation and Histone Deacetylation. BMC Genom. 2006, 7, 181. [Google Scholar] [CrossRef] [Green Version]
- Bárcena-Varela, M.; Caruso, S.; Llerena, S.; Álvarez-Sola, G.; Uriarte, I.; Latasa, U.M.; Urtasun, R.; Rebouissou, S.; Alvarez, L.; Jimenez, M.; et al. Dual Targeting of Histone Methyltransferase G9a and DNA-Methyltransferase 1 for the Treatment of Experimental Hepatocellular Carcinoma. Hepatology 2019, 69, 587–603. [Google Scholar] [CrossRef]
- Hsiao, L.-L.; Dangond, F.; Yoshida, T.; Hong, R.; Jensen, R.V.; Misra, J.; Dillon, W.; Lee, K.F.; Clark, K.E.; Haverty, P.; et al. A Compendium of Gene Expression in Normal Human Tissues. Physiol. Genom. 2001, 7, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Morgan, H.D.; Santos, F.; Green, K.; Dean, W.; Reik, W. Epigenetic Reprogramming in Mammals. Hum. Mol. Genet 2005, 14, R47–R58. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Barrena, M.G.; Arechederra, M.; Colyn, L.; Berasain, C.; Avila, M.A. Epigenetics in Hepatocellular Carcinoma Development and Therapy: The Tip of the Iceberg. Jhep Rep. 2020, 2, 100167. [Google Scholar] [CrossRef] [PubMed]
- Eden, A.; Gaudet, F.; Waghmare, A.; Jaenisch, R. Chromosomal Instability and Tumors Promoted by DNA Hypomethylation. Science 2003, 300, 455. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Meza, G.; Felden, J.; Gonzalez-Kozlova, E.E.; Garcia-Lezana, T.; Peix, J.; Portela, A.; Craig, A.J.; Sayols, S.; Schwartz, M.; Losic, B.; et al. DNA Methylation Profiling of Human Hepatocarcinogenesis. Hepatology 2021, 74, 183–199. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.; Zhou, B.; Liu, L.; Yang, F.; Conran, C.; Ji, Y.; Hou, J.; Jiang, D. Molecular Pattern of LncRNAs in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. Cr. 2019, 38, 198. [Google Scholar] [CrossRef] [Green Version]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive Analysis of Normal Adjacent to Tumor Transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [Green Version]
- Esposti, D.D.; Hernandez-Vargas, H.; Voegele, C.; Fernandez-Jimenez, N.; Forey, N.; Bancel, B.; Calvez-Kelm, F.L.; McKay, J.; Merle, P.; Herceg, Z. Identification of Novel Long Non-Coding RNAs Deregulated in Hepatocellular Carcinoma Using RNA-Sequencing. Oncotarget 2016, 7, 31862–31877. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Shi, H.; Wang, X.; Wang, B.; Qu, Q.; Geng, H.; Sun, H. Identification of Diagnostic Long Non-Coding RNA Biomarkers in Patients with Hepatocellular Carcinoma. Mol. Med. Rep. 2019, 20, 1121–1130. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Zuo, Q.; Hu, B.; Jin, H.; Wang, C.; Cheng, Z.; Deng, X.; Yang, C.; Ruan, H.; Yu, C.; et al. A Novel, Liver-Specific Long Noncoding RNA LINC01093 Suppresses HCC Progression by Interaction with IGF2BP1 to Facilitate Decay of GLI1 MRNA. Cancer Lett. 2019, 450, 98–109. [Google Scholar] [CrossRef]
- Burenina, O.Y.; Lazarevich, N.L.; Kustova, I.F.; Shavochkina, D.A.; Moroz, E.A.; Kudashkin, N.E.; Patyutko, Y.I.; Metelin, A.V.; Kim, E.F.; Skvortsov, D.A.; et al. Panel of Potential LncRNA Biomarkers Can Distinguish Various Types of Liver Malignant and Benign Tumors. J. Cancer Res. Clin. 2021, 147, 49–59. [Google Scholar] [CrossRef]
- Zhao, B.; Ke, K.; Wang, Y.; Wang, F.; Shi, Y.; Zheng, X.; Yang, X.; Liu, X.; Liu, J. HIF-1α and HDAC1 Mediated Regulation of FAM99A-MiR92a Signaling Contributes to Hypoxia Induced HCC Metastasis. Signal. Transduct. Target Ther. 2020, 5, 118. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.; Liu, S.; Ma, X.; Tan, C.; Wei, L.; Sheng, Y.; Song, Y.; Zeng, X.; Huang, D.; Qiu, X. A Liver-Specific LncRNA, FAM99B, Suppresses Hepatocellular Carcinoma Progression through Inhibition of Cell Proliferation, Migration, and Invasion. J. Cancer Res. Clin. 2019, 145, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- Jing, G.; Zheng, X.; Ji, X. LncRNA HAND2-AS1 Overexpression Inhibits Cancer Cell Proliferation in Hepatocellular Carcinoma by Downregulating RUNX2 Expression. J. Clin. Lab Anal. 2021, 35, e23717. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.-Q.; Li, Z.-H.; Zhang, H. Long Noncoding RNA HAND2-AS1 Reduced the Viability of Hepatocellular Carcinoma via Targeting MicroRNA-300/SOCS5 Axis. Hepatob. Pancreat. Dis. 2020, 19, 567–574. [Google Scholar] [CrossRef]
- Lin, B.; He, H.; Zhang, Q.; Zhang, J.; Xu, L.; Zhou, L.; Zheng, S.; Wu, L. Long Non-Coding RNA00844 Inhibits MAPK Signaling to Suppress the Progression of Hepatocellular Carcinoma by Targeting AZGP1. Ann. Transl. Med. 2020, 8, 1365. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Huang, K.; Zhang, Q.; Ye, S.; Zhong, Z.; Zeng, C.; Peng, G.; Li, L.; Ye, Q. LINC00844 Promotes Proliferation and Migration of Hepatocellular Carcinoma by Regulating NDRG1 Expression. PeerJ 2020, 8, e8394. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.F.; Zhang, Y.N.; Song, S.S.; Hu, Z.M.; He, X.L.; Pan, H.Y.; Zhang, C.W.; Wang, H.J.; Li, W.F.; Mou, X.Z. Decreased Expression of GBA3 Correlates with a Poor Prognosis in Hepatocellular Carcinoma Patients. Neoplasma 2020, 67, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zheng, Q.; Chu, Q.; Zhu, H. HAND2-AS1: A Functional Cancer-Related Long Non-Coding RNA. Biomed. Pharm. 2021, 137, 111317. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, X.; Wang, L.; Zhu, X.; Xia, Z.; Xu, L.; Xu, J. FENDRR Suppresses Cervical Cancer Proliferation and Invasion by Targeting MiR-15a/b-5p and Regulating TUBA1A Expression. Cancer Cell Int. 2020, 20, 152. [Google Scholar] [CrossRef]
- Zhang, M.-Y.; Zhang, Z.-L.; Cui, H.-X.; Wang, R.-K.; Fu, L. Long Non-Coding RNA FENDRR Inhibits NSCLC Cell Growth and Aggressiveness by Sponging MiR. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8324–8332. [Google Scholar] [CrossRef]
- Liu, L.; Yue, H.; Liu, Q.; Yuan, J.; Li, J.; Wei, G.; Chen, X.; Lu, Y.; Guo, M.; Luo, J.; et al. LncRNA MT1JP Functions as a Tumor Suppressor by Interacting with TIAR to Modulate the P53 Pathway. Oncotarget 2016, 7, 15787–15800. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Li, S. Long Non-Coding RNA MT1JP Exerts Anti-Cancer Effects in Breast Cancer Cells by Regulating MiR-92-3p. Gen. Physiol. Biophys. 2020, 39, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Esteller, M.; Rountree, M.R.; Bachman, K.E.; Schuebel, K.; Herman, J.G. Aberrant Patterns of DNA Methylation, Chromatin Formation and Gene Expression in Cancer. Hum. Mol. Genet. 2001, 10, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lv, S.; Zhong, J. Identification of LncRNA FAM99A Gene as a Prognostic Biomarker of Hepatocellular Carcinoma; Research Square: Durham, NC, USA, 2020. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique Features of Long Non-Coding RNA Biogenesis and Function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Guo, T.; Wang, H.; Liu, P.; Xiao, Y.; Wu, P.; Wang, Y.; Chen, B.; Zhao, Q.; Liu, Z.; Liu, Q. SNHG6 Acts as a Genome-Wide Hypomethylation Trigger via Coupling of MiR-1297-Mediated S-Adenosylmethionine-Dependent Positive Feedback Loops. Cancer Res. 2018, 78, 3849–3864. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Recalde, M.; Gárate-Rascón, M.; Herranz, J.M.; Elizalde, M.; Azkona, M.; Unfried, J.P.; Boix, L.; Reig, M.; Sangro, B.; Fernández-Barrena, M.G.; et al. DNA Methylation Regulates a Set of Long Non-Coding RNAs Compromising Hepatic Identity during Hepatocarcinogenesis. Cancers 2022, 14, 2048. https://doi.org/10.3390/cancers14092048
Recalde M, Gárate-Rascón M, Herranz JM, Elizalde M, Azkona M, Unfried JP, Boix L, Reig M, Sangro B, Fernández-Barrena MG, et al. DNA Methylation Regulates a Set of Long Non-Coding RNAs Compromising Hepatic Identity during Hepatocarcinogenesis. Cancers. 2022; 14(9):2048. https://doi.org/10.3390/cancers14092048
Chicago/Turabian StyleRecalde, Miriam, María Gárate-Rascón, José María Herranz, María Elizalde, María Azkona, Juan P. Unfried, Loreto Boix, María Reig, Bruno Sangro, Maite G. Fernández-Barrena, and et al. 2022. "DNA Methylation Regulates a Set of Long Non-Coding RNAs Compromising Hepatic Identity during Hepatocarcinogenesis" Cancers 14, no. 9: 2048. https://doi.org/10.3390/cancers14092048
APA StyleRecalde, M., Gárate-Rascón, M., Herranz, J. M., Elizalde, M., Azkona, M., Unfried, J. P., Boix, L., Reig, M., Sangro, B., Fernández-Barrena, M. G., Fortes, P., Ávila, M. A., Berasain, C., & Arechederra, M. (2022). DNA Methylation Regulates a Set of Long Non-Coding RNAs Compromising Hepatic Identity during Hepatocarcinogenesis. Cancers, 14(9), 2048. https://doi.org/10.3390/cancers14092048