Stratifying Risk for Pancreatic Cancer by Multiplexed Blood Test
Abstract
:Simple Summary
Abstract
1. Introduction
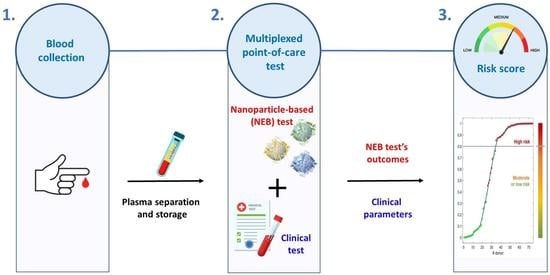
2. Materials and Methods
2.1. Patients’ Enrollment and Inclusion Criteria
2.2. Gold Nanoparticle-Enabled Blood Test
2.3. Discriminant Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Gallo, M.; Adinolfi, V.; Morviducci, L.; Acquati, S.; Tuveri, E.; Ferrari, P.; Zatelli, M.; Faggiano, A.; Argentiero, A.; Natalicchio, A. Early prediction of pancreatic cancer from new-onset diabetes: An associazione Italiana oncologia medica (AIOM)/associazione medici diabetologi (AMD)/Società Italiana endocrinologia (SIE)/Società Italiana farmacologia (SIF) multidisciplinary consensus position paper. ESMO Open 2021, 6, 100155. [Google Scholar] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Bowen, H.; Huang, H.; Zhang, S.; Zhang, D.; Shi, Q.; Liu, J.; Guo, J. Artificial intelligence in pancreatic cancer. Theranostics 2022, 12, 6931. [Google Scholar]
- Murakami, M.; Nagai, Y.; Tenjin, A.; Tanaka, Y. Proposed cut-off value of CA19-9 for detecting pancreatic cancer in patients with diabetes: A case-control study. Endocr. J. 2018, 65, 639–643. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Hruban, R.H.; Canto, M.I.; Goggins, M.; Schulick, R.; Klein, A.P. Update on familial pancreatic cancer. Adv. Surg. 2010, 44, 293–311. [Google Scholar] [CrossRef]
- Iodice, S.; Gandini, S.; Maisonneuve, P.; Lowenfels, A.B. Tobacco and the risk of pancreatic cancer: A review and meta-analysis. Langenbeck’s Arch. Surg. 2008, 393, 535–545. [Google Scholar] [CrossRef]
- Raimondi, S.; Lowenfels, A.B.; Morselli-Labate, A.M.; Maisonneuve, P.; Pezzilli, R. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract. Res. Clin. Gastroenterol. 2010, 24, 349–358. [Google Scholar] [CrossRef]
- Bosetti, C.; Rosato, V.; Li, D.; Silverman, D.; Petersen, G.; Bracci, P.; Neale, R.; Muscat, J.; Anderson, K.; Gallinger, S. Diabetes, antidiabetic medications, and pancreatic cancer risk: An analysis from the International Pancreatic Cancer Case-Control Consortium. Ann. Oncol. 2014, 25, 2065–2072. [Google Scholar] [CrossRef]
- Lynch, H.T.; Deters, C.A.; Snyder, C.L.; Lynch, J.F.; Villeneuve, P.; Silberstein, J.; Martin, H.; Narod, S.A.; Brand, R.E. BRCA1 and pancreatic cancer: Pedigree findings and their causal relationships. Cancer Genet. Cytogenet. 2005, 158, 119–125. [Google Scholar] [CrossRef]
- Iqbal, J.; Ragone, A.; Lubinski, J.; Lynch, H.; Moller, P.; Ghadirian, P.; Foulkes, W.; Armel, S.; Eisen, A.; Neuhausen, S. The incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br. J. Cancer 2012, 107, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Vanek, P.; Urban, O.; Zoundjiekpon, V.; Falt, P. Current Screening Strategies for Pancreatic Cancer. Biomedicines 2022, 10, 2056. [Google Scholar] [CrossRef] [PubMed]
- George, B.; Kent, M.; Surinach, A.; Lamarre, N.; Cockrum, P. The association of real-world CA 19-9 level monitoring patterns and clinical outcomes among patients with metastatic pancreatic ductal adenocarcinoma. Front. Oncol. 2021, 11, 754687. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846. [Google Scholar] [CrossRef]
- Azizian, A.; Rühlmann, F.; Krause, T.; Bernhardt, M.; Jo, P.; König, A.; Kleiß, M.; Leha, A.; Ghadimi, M.; Gaedcke, J. CA19-9 for detecting recurrence of pancreatic cancer. Sci. Rep. 2020, 10, 1332. [Google Scholar] [CrossRef]
- Kunovsky, L.; Tesarikova, P.; Kala, Z.; Kroupa, R.; Kysela, P.; Dolina, J.; Trna, J. The use of biomarkers in early diagnostics of pancreatic cancer. Can. J. Gastroenterol. Hepatol. 2018, 2018, 5389820. [Google Scholar] [CrossRef]
- Eid, M.; Karousi, P.; Kunovský, L.; Tuček, Š.; Brančíková, D.; Kala, Z.; Slabý, O.; Mayer, J.; Kontos, C.K.; Trna, J. The role of circulating MicroRNAs in patients with early-stage pancreatic adenocarcinoma. Biomedicines 2021, 9, 1468. [Google Scholar] [CrossRef]
- Wolrab, D.; Jirásko, R.; Cífková, E.; Höring, M.; Mei, D.; Chocholoušková, M.; Peterka, O.; Idkowiak, J.; Hrnčiarová, T.; Kuchař, L. Lipidomic profiling of human serum enables detection of pancreatic cancer. Nat. Commun. 2022, 13, 124. [Google Scholar] [CrossRef]
- Wei, L.; Yao, K.; Gan, S.; Suo, Z. Clinical utilization of serum-or plasma-based miRNAs as early detection biomarkers for pancreatic cancer: A meta-analysis up to now. Medicine 2018, 97, e12132. [Google Scholar] [CrossRef]
- Shu, X.; Zheng, W.; Yu, D.; Li, H.L.; Lan, Q.; Yang, G.; Cai, H.; Ma, X.; Rothman, N.; Gao, Y.T. P rospective metabolomics study identifies potential novel blood metabolites associated with pancreatic cancer risk. Int. J. Cancer 2018, 143, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- Caputo, D.; Digiacomo, L.; Cascone, C.; Pozzi, D.; Palchetti, S.; Di Santo, R.; Quagliarini, E.; Coppola, R.; Mahmoudi, M.; Caracciolo, G. Synergistic analysis of protein corona and haemoglobin levels detects pancreatic cancer. Cancers 2020, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.; Metzger, P.; Gerum, S.; Mayerle, J.; Schneider, G.; Belka, C.; Schnurr, M.; Lauber, K. Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches. Radiat. Oncol. 2019, 14, 141. [Google Scholar] [CrossRef]
- Quagliarini, E.; Digiacomo, L.; Renzi, S.; Pozzi, D.; Caracciolo, G. A decade of the liposome-protein corona: Lessons learned and future breakthroughs in theranostics. Nano Today 2022, 47, 101657. [Google Scholar] [CrossRef]
- Hajipour, M.J.; Laurent, S.; Aghaie, A.; Rezaee, F.; Mahmoudi, M. Personalized protein coronas: A “key” factor at the nanobiointerface. Biomater. Sci. 2014, 2, 1210–1221. [Google Scholar] [CrossRef]
- Corbo, C.; Molinaro, R.; Tabatabaei, M.; Farokhzad, O.C.; Mahmoudi, M. Personalized protein corona on nanoparticles and its clinical implications. Biomater. Sci. 2017, 5, 378–387. [Google Scholar] [CrossRef]
- Caracciolo, G.; Safavi-Sohi, R.; Malekzadeh, R.; Poustchi, H.; Vasighi, M.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Hajipour, M.; Di Domenico, M. Disease-specific protein corona sensor arrays may have disease detection capacity. Nanoscale Horiz. 2019, 4, 1063–1076. [Google Scholar] [CrossRef]
- Di Santo, R.; Digiacomo, L.; Quagliarini, E.; Capriotti, A.L.; Laganà, A.; Zenezini Chiozzi, R.; Caputo, D.; Cascone, C.; Coppola, R.; Pozzi, D. Personalized graphene oxide-protein corona in the human plasma of pancreatic cancer patients. Front. Bioeng. Biotechnol. 2020, 8, 491. [Google Scholar] [CrossRef]
- Caputo, D.; Caracciolo, G. Nanoparticle-enabled blood tests for early detection of pancreatic ductal adenocarcinoma. Cancer Lett. 2020, 470, 191–196. [Google Scholar] [CrossRef]
- Quagliarini, E.; Caputo, D.; Cammarata, R.; Caracciolo, G.; Pozzi, D. Coupling magnetic levitation of graphene oxide–protein complexes with blood levels of glucose for early detection of pancreatic adenocarcinoma. Cancer Nanotechnol. 2023, 14, 16. [Google Scholar] [CrossRef]
- Digiacomo, L.; Caputo, D.; Coppola, R.; Cascone, C.; Giulimondi, F.; Palchetti, S.; Pozzi, D.; Caracciolo, G. Efficient pancreatic cancer detection through personalized protein corona of gold nanoparticles. Biointerphases 2021, 16, 011010. [Google Scholar] [CrossRef]
- Caracciolo, G.; Pozzi, D.; Palchetti, S.; Digiacomo, L.; Caputo, D.; Coppola, R. A Method to Assist in the Early Diagnosis of Pancreatic Adenocarcinoma. U.S. Patent Application 17/629,091, 28 July 2022. [Google Scholar]
- Xu, S.-S.; Li, S.; Xu, H.-X.; Li, H.; Wu, C.-T.; Wang, W.-Q.; Gao, H.-L.; Jiang, W.; Zhang, W.-H.; Li, T.-J. Haemoglobin, albumin, lymphocyte and platelet predicts postoperative survival in pancreatic cancer. World J. Gastroenterol. 2020, 26, 828. [Google Scholar] [CrossRef] [PubMed]
- Caputo, D.; Coppola, A.; Quagliarini, E.; Di Santo, R.; Capriotti, A.L.; Cammarata, R.; Laganà, A.; Papi, M.; Digiacomo, L.; Coppola, R. Multiplexed Detection of Pancreatic Cancer by Combining a Nanoparticle-Enabled Blood Test and Plasma Levels of Acute-Phase Proteins. Cancers 2022, 14, 4658. [Google Scholar] [CrossRef] [PubMed]
- Digiacomo, L.; Palchetti, S.; Giulimondi, F.; Pozzi, D.; Chiozzi, R.Z.; Capriotti, A.L.; Laganà, A.; Caracciolo, G. The biomolecular corona of gold nanoparticles in a controlled microfluidic environment. Lab A Chip 2019, 19, 2557–2567. [Google Scholar] [CrossRef]
- Min, L.; Ziyu, D.; Xiaofei, Z.; Shunhe, X.; Bolin, W. Analysis of levels and Clinical value of CA19-9, NLR and SIRI in patients with Pancreatic Cancer with different Clinical Features. Cell. Mol. Biol. 2021, 67, 302–308. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Li, H.; Wu, Y.; Zhang, H.; Chen, W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 11683. [Google Scholar]


| NOP (N = 27) | PDAC (N = 47) | p-Value | |
|---|---|---|---|
| Albumin (g/mL) | 3.62 ± 0.57 | 3.06 ± 0.65 | 0.0003 |
| WBC (103/μL) | 7.28 ± 1.67 | 7.87 ± 3.54 | 0.4186 |
| Neutrophils (103/μL) | 4.46 ± 1.23 | 5.51 ± 3.27 | 0.1134 |
| Lymphocytes (103/μL) | 2.07 ± 0.64 | 1.64 ± 0.65 | 0.0075 |
| Platelets (103/μL) | 253 ± 92.7 | 208 ± 68.9 | 0.0182 |
| Hemoglobin (g/dL) | 14.3 ± 1.76 | 11.9 ± 2.36 | <0.0001 |
| NLR | 2.33 ± 0.95 | 4.12 ± 4.72 | 0.0564 |
| dNLR | 1.66 ± 0.55 | 2.61 ± 2.09 | 0.0235 |
| PLR | 134 ± 56.9 | 143 ± 62.7 | 0.5304 |
| Glucose (mg/dL) | 98.1 ± 12.9 | 131 ± 49.6 | 0.0011 |
| Total bilirubin (mg/dL) | 0.80 ± 0.62 | 2.31 ± 3.40 | 0.0249 |
| Direct bilirubin (mg/dL) | 0.16 ± 0.08 | 1.63 ± 2.85 | 0.0096 |
| GOT (U/L) | 19.5 ± 8.49 | 49.6 ± 56.5 | 0.0078 |
| NEB1 | 0.22 ± 0.04 | 0.31 ± 0.08 | <0.0001 |
| NEB2 | 0.21 ± 0.05 | 0.21 ± 0.05 | 0.5040 |
| NEB3 | 0.34 ± 0.09 | 0.28 ± 0.06 | 0.0007 |
| NEB4 | 0.24 ± 0.08 | 0.20 ± 0.06 | 0.0231 |
| NOP (N = 27) | PDAC (N = 47) | |
|---|---|---|
| Age, median (range) | 58.5 (23–84) | 71 (47–83) |
| Sex, N (%) | ||
| Male | 13 (48.1%) | 23 (62.5%) |
| Female | 14 (51.9%) | 24 (37.5%) |
| Comorbidity, N (%) | ||
| Cardiac | 2 (7%) | 15 (31.9%) |
| Pulmonary | 1 (3.7%) | 4 (8.5%) |
| Diabetes mellitus | 1 (3.7%) | 9 (19.1%) |
| Hypertension | 3 (11.1%) | 2 (4.2%) |
| None | 15 (55.5%) | 1 (2.1%) |
| Smoking status, N (%) | ||
| Never | 13 (48.2%) | 12 (25.5%) |
| Ex-smoker | 7 (25.9%) | 16 (34%) |
| Current | 7 (25.9%) | 19 (40.5%) |
| CA 19.9 (UI/l), N (%) | ||
| >37.00 | 0 | 36 (76.5%) |
| <37.00 | 5 (18.5%) | 11 (23.5%) |
| Missing data | 22 (81.5%) | 0 |
| TNM stage, N (%) | ||
| I | NA | 2 (4.4%) |
| II | NA | 8 (17%) |
| III | NA | 30 (63.8%) |
| IV | NA | 7 (14.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Digiacomo, L.; Quagliarini, E.; Pozzi, D.; Coppola, R.; Caracciolo, G.; Caputo, D. Stratifying Risk for Pancreatic Cancer by Multiplexed Blood Test. Cancers 2023, 15, 2983. https://doi.org/10.3390/cancers15112983
Digiacomo L, Quagliarini E, Pozzi D, Coppola R, Caracciolo G, Caputo D. Stratifying Risk for Pancreatic Cancer by Multiplexed Blood Test. Cancers. 2023; 15(11):2983. https://doi.org/10.3390/cancers15112983
Chicago/Turabian StyleDigiacomo, Luca, Erica Quagliarini, Daniela Pozzi, Roberto Coppola, Giulio Caracciolo, and Damiano Caputo. 2023. "Stratifying Risk for Pancreatic Cancer by Multiplexed Blood Test" Cancers 15, no. 11: 2983. https://doi.org/10.3390/cancers15112983
APA StyleDigiacomo, L., Quagliarini, E., Pozzi, D., Coppola, R., Caracciolo, G., & Caputo, D. (2023). Stratifying Risk for Pancreatic Cancer by Multiplexed Blood Test. Cancers, 15(11), 2983. https://doi.org/10.3390/cancers15112983