MicroRNAs as Predictive Biomarkers in Patients with Colorectal Cancer Receiving Chemotherapy or Chemoradiotherapy: A Narrative Literature Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
3. Results
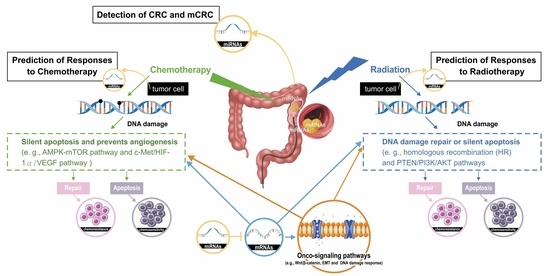
3.1. MiRs Associated with the Detection of CRC and mCRC
3.2. MiRs Associated with the Prediction of Responses to Chemotherapy in CRC
3.3. MiRs Associated with the Prediction of Responses to Radiotherapy in CRC
| Family | MiRs | Verified Targets in CRC or Other Cancers | Sample Source | Target for miRNA | Ref. |
|---|---|---|---|---|---|
| miR-17 | miR-17, miR-18a/b, miR-106a/b, miR-20a/b, miR-93 | MiR-106b induces cell radioresistance by targeting the PTEN/PI3K/AKT pathways and p21 in CRC. MiR-93 acts as a specific exosomal cargo that increases radioresistance. Inhibition of miR-93 suppressed radioresistance. | Tumor tissue Tumor tissue | PTEN/PI3K/AKT pathways and p21 FOXA1 BTG3 | [130] [131] [133] |
| miR-19 | miR-19a, miR-19b-1, miR-19b-2 | The low miR-19b expression levels of patients with LARC had markedly longer OS and DFS. MiR-19b induces radioresistance, and the patients with higher miR-19b expression levels had a shorter survival time. | Tumor tissue | FBXW7 | [135] [136] |
| miR-96 | miR-96 | miR-96-5p induced radioresistance is upregulated in rectal cancer cells through the inhibition of GPC3 and abnormal triggering of the canonical Wnt signaling pathway. | Tumor tissue | GPC3 | [134] |
| miR-103 | miR-103a/b, miR-107 | MiR-107 induces chemoresistance. Hsa-mir-107 and WDFY3-AS2 may serve as prognostic biomarkers in RC. | Tumor tissue | Through the CAB39–AMPK–mTOR pathway | [89] [91] |
| miR-221 | miR-221, miR-222 | MiR-222 induces radiation resistance. | Serum | PTEN. | [104] |
3.4. MiRs Associated with the Prediction of Responses to Chemoradiotherapy in CRC
3.5. Current Undergoing Clinical Trials for miRs
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Cancer Society Cancer Statistics 2021 Report. J. Nucl. Med. 2021, 62, 12N.
- Qaderi, S.M.; Galjart, B.; Verhoef, C.; Slooter, G.D.; Koopman, M.; Verhoeven, R.H.A.; de Wilt, J.H.W.; van Erning, F.N. Disease recurrence after colorectal cancer surgery in the modern era: A population-based study. Int. J. Colorectal. Dis. 2021, 36, 2399–2410. [Google Scholar] [CrossRef]
- Andre, T.; Boni, C.; Mounedji-Boudiaf, L.; Navarro, M.; Tabernero, J.; Hickish, T.; Topham, C.; Zaninelli, M.; Clingan, P.; Bridgewater, J.; et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N. Engl. J. Med. 2004, 350, 2343–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Twelves, C.; Wong, A.; Nowacki, M.P.; Abt, M.; Burris, H., 3rd; Carrato, A.; Cassidy, J.; Cervantes, A.; Fagerberg, J.; Georgoulias, V.; et al. Capecitabine as adjuvant treatment for stage III colon cancer. N. Engl. J. Med. 2005, 352, 2696–2704. [Google Scholar] [CrossRef]
- Huang, M.Y.; Huang, C.M.; Tsai, H.L.; Huang, C.W.; Hsieh, H.M.; Yeh, Y.S.; Wu, J.Y.; Wang, W.M.; Wang, J.Y. Comparison of adjuvant FOLFOX4 chemotherapy and oral UFUR/LV following adjuvant FOLFOX4 chemotherapy in patients with stage III colon cancer subsequent to radical resection. Oncol. Lett. 2017, 14, 6754–6762. [Google Scholar] [CrossRef]
- Goldberg, R.M.; Sargent, D.J.; Morton, R.F.; Fuchs, C.S.; Ramanathan, R.K.; Williamson, S.K.; Findlay, B.P.; Pitot, H.C.; Alberts, S.R. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J. Clin. Oncol. 2004, 22, 23–30. [Google Scholar] [CrossRef]
- Yang, A.D.; Fan, F.; Camp, E.R.; van Buren, G.; Liu, W.; Somcio, R.; Gray, M.J.; Cheng, H.; Hoff, P.M.; Ellis, L.M. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin. Cancer Res. 2006, 12, 4147–4153. [Google Scholar] [CrossRef] [Green Version]
- Andre, T.; Boni, C.; Navarro, M.; Tabernero, J.; Hickish, T.; Topham, C.; Bonetti, A.; Clingan, P.; Bridgewater, J.; Rivera, F.; et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J. Clin. Oncol. 2009, 27, 3109–3116. [Google Scholar] [CrossRef] [Green Version]
- Su, M.W.; Chang, C.K.; Lin, C.W.; Chu, H.W.; Tsai, T.N.; Su, W.C.; Chen, Y.C.; Chang, T.K.; Huang, C.W.; Tsai, H.L.; et al. Genomic and Metabolomic Landscape of Right-Sided and Left-Sided Colorectal Cancer: Potential Preventive Biomarkers. Cells 2022, 11, 527. [Google Scholar] [CrossRef]
- Chen, H.H.; Ke, T.W.; Huang, C.W.; Jiang, J.K.; Chen, C.C.; Hsieh, Y.Y.; Teng, H.W.; Lin, B.W.; Liang, Y.H.; Su, Y.L.; et al. Taiwan Society of Colon and Rectal Surgeons Consensus on mCRC Treatment. Front. Oncol. 2021, 11, 764912. [Google Scholar] [CrossRef] [PubMed]
- Tsujii, M. Search for novel target molecules for the effective treatment or prevention of colorectal cancer. Digestion 2012, 85, 99–102. [Google Scholar] [CrossRef]
- Karoui, M.; Rullier, A.; Piessen, G.; Legoux, J.L.; Barbier, E.; De Chaisemartin, C.; Lecaille, C.; Bouche, O.; Ammarguellat, H.; Brunetti, F.; et al. Perioperative FOLFOX 4 Versus FOLFOX 4 Plus Cetuximab Versus Immediate Surgery for High-Risk Stage II and III Colon Cancers: A Phase II Multicenter Randomized Controlled Trial (PRODIGE 22). Ann. Surg. 2020, 271, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Cremolini, C.; Antoniotti, C.; Lonardi, S.; Bergamo, F.; Cortesi, E.; Tomasello, G.; Moretto, R.; Ronzoni, M.; Racca, P.; Loupakis, F.; et al. Primary tumor sidedness and benefit from FOLFOXIRI plus bevacizumab as initial therapy for metastatic colorectal cancer. Retrospective analysis of the TRIBE trial by GONO. Ann. Oncol. 2018, 29, 1528–1534. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- De Felice, F.; Musio, D.; Benevento, I.; Magnante, A.; Bulzonetti, N.; Rossella, C.; Tombolini, V. Influence of Organ Invasion in Clinical Outcomes for Locally Advanced Rectal Cancer. Anticancer Res. 2016, 36, 5443–5447. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Yu, X.; Xiao, W.W.; Wang, Q.X.; Zhou, W.H.; Zeng, Z.F.; Ding, P.R.; Li, L.R.; Gao, Y.H. Neoadjuvant chemoradiotherapy followed by surgery in patients with unresectable locally advanced colon cancer: A prospective observational study. OncoTargets Ther. 2018, 11, 409–418. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.Y.; Huang, C.W.; Wang, J.Y. Surgical treatment following neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Kaohsiung J. Med. Sci. 2020, 36, 152–159. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.M.; Huang, M.Y.; Tsai, H.L.; Huang, C.W.; Ma, C.J.; Yeh, Y.S.; Juo, S.H.; Huang, C.J.; Wang, J.Y. An observational study of extending FOLFOX chemotherapy, lengthening the interval between radiotherapy and surgery, and enhancing pathological complete response rates in rectal cancer patients following preoperative chemoradiotherapy. Therap. Adv. Gastroenterol. 2016, 9, 702–712. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.W.; Su, W.C.; Yin, T.C.; Chen, P.J.; Chang, T.K.; Chen, Y.C.; Li, C.C.; Hsieh, Y.C.; Tsai, H.L.; Wang, J.Y. Time interval between the completion of radiotherapy and robotic-assisted surgery among patients with stage I-III rectal cancer undergoing preoperative chemoradiotherapy. PLoS ONE 2020, 15, e0240742. [Google Scholar] [CrossRef]
- Hu, X.; Li, Y.Q.; Ma, X.J.; Zhang, L.; Cai, S.J.; Peng, J.J. Adjuvant chemotherapy for rectal cancer with complete pathological response (pCR) may not be necessary: A pooled analysis of 5491 patients. Cancer Cell Int. 2019, 19, 127. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.Y.; Huang, J.J.; Huang, C.M.; Lin, C.H.; Tsai, H.L.; Huang, C.W.; Chai, C.Y.; Lin, C.Y.; Wang, J.Y. Relationship Between Expression of Proteins ERCC1, ERCC2, and XRCC1 and Clinical Outcomes in Patients with Rectal Cancer Treated with FOLFOX-Based Preoperative Chemoradiotherapy. World J. Surg. 2017, 41, 2884–2897. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.K.; Mukherjee, S. Assimilating Epigenetics and Transcriptomics for the Identification of Prognostic Novel Biomarkers and Imminent Targets in Colorectal Carcinoma with Therapeutic Potential. Curr. Mol. Med. 2022. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.; Hernandez-Illan, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Li Destri, G.; Basile, G.; Agodi, A. Epigenetic Biomarkers in Colorectal Cancer Patients Receiving Adjuvant or Neoadjuvant Therapy: A Systematic Review of Epidemiological Studies. Int. J. Mol. Sci. 2019, 20, 3842. [Google Scholar] [CrossRef] [Green Version]
- Schraa, S.J.; van Rooijen, K.L.; Koopman, M.; Vink, G.R.; Fijneman, R.J.A. Cell-Free Circulating (Tumor) DNA before Surgery as a Prognostic Factor in Non-Metastatic Colorectal Cancer: A Systematic Review. Cancers 2022, 14, 2218. [Google Scholar] [CrossRef]
- Bjorkman, K.; Jalkanen, S.; Salmi, M.; Mustonen, H.; Kaprio, T.; Kekki, H.; Pettersson, K.; Bockelman, C.; Haglund, C. A prognostic model for colorectal cancer based on CEA and a 48-multiplex serum biomarker panel. Sci. Rep. 2021, 11, 4287. [Google Scholar] [CrossRef]
- Sun, W.; Huang, T.; Li, G.; Shen, W.; Zhu, J.; Jin, Q.; Zhao, J.; Jia, C.; Zhang, Z. The advantage of circulating tumor cells over serum carcinoembryonic antigen for predicting treatment responses in rectal cancer. Future Oncol. 2013, 9, 1489–1500. [Google Scholar] [CrossRef]
- Huang, C.W.; Chen, Y.T.; Tsai, H.L.; Yeh, Y.S.; Su, W.C.; Ma, C.J.; Tsai, T.N.; Wang, J.Y. EGFR expression in patients with stage III colorectal cancer after adjuvant chemotherapy and on cancer cell function. Oncotarget 2017, 8, 114663–114676. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.J.; Ren, S.N.; Liu, Y.T.; Yan, H.W.; Chen, X.B. Targeting EGFR sensitizes 5-Fu-resistant colon cancer cells through modification of the lncRNA-FGD5-AS1-miR-330-3p-Hexokinase 2 axis. Mol. Ther. Oncol. 2021, 23, 14–25. [Google Scholar] [CrossRef]
- Huang, M.Y.; Tsai, H.L.; Huang, J.J.; Wang, J.Y. Clinical Implications and Future Perspectives of Circulating Tumor Cells and Biomarkers in Clinical Outcomes of Colorectal Cancer. Transl. Oncol. 2016, 9, 340–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.Y.; Uen, Y.H.; Tsai, H.L.; Chuang, S.C.; Hou, M.F.; Wu, D.C.; Juo, S.H.; Lin, S.R.; Wang, J.Y. Molecular detection of persistent postoperative circulating tumour cells in stages II and III colon cancer patients via multiple blood sampling: Prognostic significance of detection for early relapse. Br. J. Cancer 2011, 104, 1178–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, M.; Zheng, M.; Xu, Y.; Ma, S.; Zhang, W.; Ju, S. CircRNAs and their regulatory roles in cancers. Mol. Med. 2021, 27, 94. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Chen, C.H.; Huang, Y.H.; Chiang, C.H.; Huang, M.Y. Biomarkers of Favorable vs. Unfavorable Responses in Locally Advanced Rectal Cancer Patients Receiving Neoadjuvant Concurrent Chemoradiotherapy. Cells 2022, 11, 1611. [Google Scholar] [CrossRef] [PubMed]
- Szeglin, B.C.; Wu, C.; Marco, M.R.; Park, H.S.; Zhang, Z.; Zhang, B.; Garcia-Aguilar, J.; Beauchamp, R.D.; Chen, X.S.; Smith, J.J. A SMAD4-modulated gene profile predicts disease-free survival in stage II and III colorectal cancer. Cancer Rep. 2022, 5, e1423. [Google Scholar] [CrossRef]
- de Assis, J.V.; Coutinho, L.A.; Oyeyemi, I.T.; Oyeyemi, O.T.; Grenfell, R. Diagnostic and therapeutic biomarkers in colorectal cancer: A review. Am. J. Cancer Res. 2022, 12, 661–680. [Google Scholar] [PubMed]
- Huang, Z.; Yang, M. Molecular Network of Colorectal Cancer and Current Therapeutic Options. Front. Oncol. 2022, 12, 852927. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Fessler, E.; Jansen, M.; De Sousa, E.M.F.; Zhao, L.; Prasetyanti, P.R.; Rodermond, H.; Kandimalla, R.; Linnekamp, J.F.; Franitza, M.; van Hooff, S.R.; et al. A multidimensional network approach reveals microRNAs as determinants of the mesenchymal colorectal cancer subtype. Oncogene 2016, 35, 6026–6037. [Google Scholar] [CrossRef] [Green Version]
- Sugai, T.; Osakabe, M.; Niinuma, T.; Sugimoto, R.; Eizuka, M.; Tanaka, Y.; Yanagawa, N.; Otsuka, K.; Sasaki, A.; Matsumoto, T.; et al. Genome-Wide Analysis of microRNA and mRNA Expression in Colorectal Intramucosal Neoplasia and Colorectal Cancer With a Microsatellite-Stable Phenotype Based on Adenoma-Carcinoma Sequences. Front. Oncol. 2022, 12, 831100. [Google Scholar] [CrossRef]
- Martens-de Kemp, S.R.; Komor, M.A.; Hegi, R.; Bolijn, A.S.; Tijssen, M.; de Groen, F.L.M.; Depla, A.; van Leerdam, M.; Meijer, G.A.; Fijneman, R.J.A.; et al. Overexpression of the miR-17-92 cluster in colorectal adenoma organoids causes a carcinoma-like gene expression signature. Neoplasia 2022, 32, 100820. [Google Scholar] [CrossRef] [PubMed]
- Paz-Cabezas, M.; Calvo-Lopez, T.; Romera-Lopez, A.; Tabas-Madrid, D.; Ogando, J.; Fernandez-Acenero, M.J.; Sastre, J.; Pascual-Montano, A.; Manes, S.; Diaz-Rubio, E.; et al. Molecular Classification of Colorectal Cancer by microRNA Profiling: Correlation with the Consensus Molecular Subtypes (CMS) and Validation of miR-30b Targets. Cancers 2022, 14, 5175. [Google Scholar] [CrossRef] [PubMed]
- Strubberg, A.M.; Madison, B.B. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis. Model Mech. 2017, 10, 197–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motoyama, K.; Inoue, H.; Takatsuno, Y.; Tanaka, F.; Mimori, K.; Uetake, H.; Sugihara, K.; Mori, M. Over- and under-expressed microRNAs in human colorectal cancer. Int. J. Oncol. 2009, 34, 1069–1075. [Google Scholar] [PubMed] [Green Version]
- Monzo, M.; Navarro, A.; Bandres, E.; Artells, R.; Moreno, I.; Gel, B.; Ibeas, R.; Moreno, J.; Martinez, F.; Diaz, T.; et al. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008, 18, 823–833. [Google Scholar] [CrossRef] [Green Version]
- Vickers, A.J.; Bianco, F.J., Jr.; Boorjian, S.; Scardino, P.T.; Eastham, J.A. Does a delay between diagnosis and radical prostatectomy increase the risk of disease recurrence? Cancer 2006, 106, 576–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schepeler, T.; Reinert, J.T.; Ostenfeld, M.S.; Christensen, L.L.; Silahtaroglu, A.N.; Dyrskjot, L.; Wiuf, C.; Sorensen, F.J.; Kruhoffer, M.; Laurberg, S.; et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008, 68, 6416–6424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwasaki, H.; Shimura, T.; Kitagawa, M.; Yamada, T.; Nishigaki, R.; Fukusada, S.; Okuda, Y.; Katano, T.; Horike, S.I.; Kataoka, H. A Novel Urinary miRNA Biomarker for Early Detection of Colorectal Cancer. Cancers 2022, 14, 461. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.P.; Tsai, H.L.; Miao, Z.F.; Huang, C.W.; Kuo, C.H.; Wu, J.Y.; Wang, W.M.; Juo, S.H.; Wang, J.Y. Development of a deregulating microRNA panel for the detection of early relapse in postoperative colorectal cancer patients. J. Transl. Med. 2016, 14, 108. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, S.J.; Netz, U.; Hallion, J.; Bishop, C.; Stephen, V.; Burton, J.; Paas, M.; Feagins, K.; Pan, J.; Rai, S.N.; et al. Circulating plasma microRNAs in colorectal neoplasia: A pilot study in assessing response to therapy. Transl. Oncol. 2021, 14, 100962. [Google Scholar] [CrossRef]
- Han, L.; Shi, W.J.; Xie, Y.B.; Zhang, Z.G. Diagnostic value of four serum exosome microRNAs panel for the detection of colorectal cancer. World J. Gastrointest. Oncol. 2021, 13, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Cekaite, L.; Eide, P.W.; Lind, G.E.; Skotheim, R.I.; Lothe, R.A. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget 2016, 7, 6476–6505. [Google Scholar] [CrossRef] [Green Version]
- Stiegelbauer, V.; Perakis, S.; Deutsch, A.; Ling, H.; Gerger, A.; Pichler, M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J. Gastroenterol. 2014, 7, 11727–11735. [Google Scholar] [CrossRef]
- Masuda, T.; Hayashi, N.; Kuroda, Y.; Ito, S.; Eguchi, H.; Mimor, K. MicroRNAs as Biomarkers in Colorectal Cancer. Cancers 2017, 9, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirafkan, N.; Mansoori, B.; Mohammadi, A.; Shomali, N.; Ghasbi, M.; Baradaran, B. MicroRNAs as novel biomarkers for colorectal cancer: New outlooks. Biomed. Pharmacother. 2018, 97, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- WaiHon, K.; Othman, N.; Hanif, E.A.M.; Nasir, S.N.; AbdRazak, N.S.; Jamal, R.; Abu, N. Cancer Sensitizing Agents for Chemotherapy; Elsevier Science: Amsterdam, The Netherlands, 2020; Volume 8, pp. 135–151. [Google Scholar]
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef]
- Wang, Y.; Jatkoe, T.; Zhang, Y.; Mutch, M.G.; Talantov, D.; Jiang, J.; McLeod, H.L.; Atkins, D. Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J. Clin. Oncol. 2004, 22, 1564–1571. [Google Scholar] [CrossRef]
- Yeh, Y.S.; Wang, H.M.; Lin, S.R.; Wang, J.Y. Prognostic and Molecular Factors in Stage II Colorectal Cancer. Genom. Med. Biomark. Health Sci. 2011, 3, 2–8. [Google Scholar] [CrossRef] [Green Version]
- Yang, I.P.; Tsai, H.L.; Hou, M.F.; Chen, K.C.; Tsai, P.C.; Huang, S.W.; Chou, W.W.; Wang, J.Y.; Juo, S.H. MicroRNA-93 inhibits tumor growth and early relapse of human colorectal cancer by affecting genes involved in the cell cycle. Carcinogenesis 2012, 33, 1522–1530. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Xu, A.; Li, J.; Fu, J.; Wang, G.; Yang, Y.; Cui, L.; Sun, J. Fecal miR-29a and miR-224 as the noninvasive biomarkers for colorectal cancer. Cancer Biomark. 2016, 16, 259–264. [Google Scholar] [CrossRef]
- Aalami, A.H.; Abdeahad, H.; Mesgari, M. Circulating miR-21 as a potential biomarker in human digestive system carcinoma: A systematic review and diagnostic meta-analysis. Biomarkers 2021, 26, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Ak, S.; Tunca, B.; Tezcan, G.; Cecener, G.; Egeli, U.; Yilmazlar, T.; Ozturk, E.; Yerci, O. MicroRNA expression patterns of tumors in early-onset colorectal cancer patients. J. Surg. Res. 2014, 191, 113–122. [Google Scholar] [CrossRef]
- Liu, K.; Li, G.; Fan, C.; Zhou, X.; Wu, B.; Li, J. Increased expression of microRNA-21and its association with chemotherapeutic response in human colorectal cancer. J. Int. Med. Res. 2011, 39, 2288–2295. [Google Scholar] [CrossRef] [Green Version]
- Menendez, P.; Padilla, D.; Villarejo, P.; Palomino, T.; Nieto, P.; Menendez, J.M.; Rodriguez-Montes, J.A. Prognostic implications of serum microRNA-21 in colorectal cancer. J. Surg. Oncol. 2013, 108, 369–373. [Google Scholar] [CrossRef]
- Yang, I.P.; Tsai, H.L.; Huang, C.W.; Huang, M.Y.; Hou, M.F.; Juo, S.H.; Wang, J.Y. The functional significance of microRNA-29c in patients with colorectal cancer: A potential circulating biomarker for predicting early relapse. PLoS ONE 2013, 8, e66842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, H.L.; Yang, I.P.; Huang, C.W.; Ma, C.J.; Kuo, C.H.; Lu, C.Y.; Juo, S.H.; Wang, J.Y. Clinical significance of microRNA-148a in patients with early relapse of stage II stage and III colorectal cancer after curative resection. Transl. Res. 2013, 162, 258–268. [Google Scholar] [CrossRef]
- Tsai, H.L.; Miao, Z.F.; Chen, Y.T.; Huang, C.W.; Yeh, Y.S.; Yang, I.P.; Wang, J.Y. miR-148a inhibits early relapsed colorectal cancers and the secretion of VEGF by indirectly targeting HIF-1alpha under non-hypoxia/hypoxia conditions. J. Cell Mol. Med. 2019, 23, 3572–3582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Xi, J.; Xu, X.; Peng, B.; Zhang, B. MiR-148a suppressed cell invasion and migration via targeting WNT10b and modulating beta-catenin signaling in cisplatin-resistant colorectal cancer cells. Biomed. Pharmacother. 2019, 109, 902–909. [Google Scholar] [CrossRef]
- Pidikova, P.; Herichova, I. miRNA Clusters with Up-Regulated Expression in Colorectal Cancer. Cancers 2021, 13, 2979. [Google Scholar] [CrossRef] [PubMed]
- Ulivi, P.; Canale, M.; Passardi, A.; Marisi, G.; Valgiusti, M.; Frassineti, G.L.; Calistri, D.; Amadori, D.; Scarpi, E. Circulating Plasma Levels of miR-20b, miR-29b and miR-155 as Predictors of Bevacizumab Efficacy in Patients with Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Liu, H.; Ning, S.; Wei, C.; Li, J.; Wei, W.; Zhang, L. The High Ratio of the Plasma miR-96/miR-99b Correlated With Poor Prognosis in Patients With Metastatic Colorectal Cancer. Front. Mol. Biosci. 2021, 8, 799060. [Google Scholar] [CrossRef]
- Hoye, E.; Fromm, B.; Bottger, P.H.M.; Domanska, D.; Torgunrud, A.; Lund-Andersen, C.; Abrahamsen, T.W.; Fretland, A.A.; Dagenborg, V.J.; Lorenz, S.; et al. A comprehensive framework for analysis of microRNA sequencing data in metastatic colorectal cancer. NAR Cancer 2022, 4, zcab051. [Google Scholar] [CrossRef]
- Lai, P.S.; Chang, W.M.; Chen, Y.Y.; Lin, Y.F.; Liao, H.F.; Chen, C.Y. Circulating microRNA-762 upregulation in colorectal cancer may be accompanied by Wnt-1/beta-catenin signaling. Cancer Biomark. 2021, 32, 111–122. [Google Scholar] [CrossRef]
- Wang, S.; Xiang, J.; Li, Z.; Lu, S.; Hu, J.; Gao, X.; Yu, L.; Wang, L.; Wang, J.; Wu, Y.; et al. A plasma microRNA panel for early detection of colorectal cancer. Int. J. Cancer 2015, 136, 152–161. [Google Scholar] [CrossRef]
- Huang, G.; Wei, B.; Chen, Z.; Wang, J.; Zhao, L.; Peng, X.; Liu, K.; Lai, Y.; Ni, L. Identification of a four-microRNA panel in serum as promising biomarker for colorectal carcinoma detection. Biomark. Med. 2020, 14, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Kohne, C.H. Successes and limitations of targeted cancer therapy in colon cancer. Prog. Tumor Res. 2014, 41, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Dahan, L.; Sadok, A.; Formento, J.L.; Seitz, J.F.; Kovacic, H. Modulation of cellular redox state underlies antagonism between oxaliplatin and cetuximab in human colorectal cancer cell lines. Br. J. Pharmacol. 2009, 158, 610–620. [Google Scholar] [CrossRef] [Green Version]
- Goh, A.M.; Coffill, C.R.; Lane, D.P. The role of mutant p53 in human cancer. J. Pathol. 2011, 223, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.K.; Yadav, V.K.; Bajaj, S.; Datta, A.; Dutta, S.K.; Bhattacharyya, M.; Bhattacharya, S.; Debnath, S.; Roy, S.; Boardman, L.A.; et al. DNA damage-induced ephrin-B2 reverse signaling promotes chemoresistance and drives EMT in colorectal carcinoma harboring mutant p53. Cell Death Differ. 2016, 23, 707–722. [Google Scholar] [CrossRef] [Green Version]
- Tanikawa, C.; Nakagawa, H.; Furukawa, Y.; Nakamura, Y.; Matsuda, K. CLCA2 as a p53-inducible senescence mediator. Neoplasia 2012, 14, 141–149. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Sun, Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene 2007, 26, 2707–2716. [Google Scholar] [CrossRef] [Green Version]
- Sadasivam, S.; Gupta, S.; Radha, V.; Batta, K.; Kundu, T.K.; Swarup, G. Caspase-1 activator Ipaf is a p53-inducible gene involved in apoptosis. Oncogene 2005, 24, 627–636. [Google Scholar] [CrossRef] [Green Version]
- Ramesh, P.; Lannagan, T.R.M.; Jackstadt, R.; Atencia Taboada, L.; Lansu, N.; Wirapati, P.; van Hooff, S.R.; Dekker, D.; Pritchard, J.; Kirov, A.B.; et al. BCL-XL is crucial for progression through the adenoma-to-carcinoma sequence of colorectal cancer. Cell Death Differ. 2021, 28, 3282–3296. [Google Scholar] [CrossRef]
- Tie, Y.; Chen, C.; Yang, Y.; Qian, Z.; Yuan, H.; Wang, H.; Tang, H.; Peng, Y.; Du, X.; Liu, B. Upregulation of let-7f-5p promotes chemotherapeutic resistance in colorectal cancer by directly repressing several pro-apoptotic proteins. Oncol. Lett. 2018, 15, 8695–8702. [Google Scholar] [CrossRef] [PubMed]
- Ge, T.; Xiang, P.; Mao, H.; Tang, S.; Zhou, J.; Zhang, Y. Inhibition of miR-96 enhances the sensitivity of colorectal cancer cells to oxaliplatin by targeting TPM1. Exp. Ther. Med. 2020, 20, 2134–2140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, J.; Li, N.; Liu, Z.; Chen, Z.; Li, Z.; Lai, Y.; Shen, L.; Gao, J. miR-34a increases the sensitivity of colorectal cancer cells to 5-fluorouracil in vitro and in vivo. Am. J. Cancer Res. 2018, 8, 280–290. [Google Scholar] [PubMed]
- Barisciano, G.; Colangelo, T.; Rosato, V.; Muccillo, L.; Taddei, M.L.; Ippolito, L.; Chiarugi, P.; Galgani, M.; Bruzzaniti, S.; Matarese, G.; et al. miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. Br. J. Cancer 2020, 122, 1354–1366. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Zhu, D.; Hou, L.; Wang, Y.; Huang, X.; Zhou, C.; Zhu, L.; Wang, Y.; Li, L.; Gu, Y.; et al. MiR-107 confers chemoresistance to colorectal cancer by targeting calcium-binding protein 39. Br. J. Cancer 2020, 122, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Tao, F.; Zhang, X.; Zhang, Y.; Sun, X.; Wu, D. Role of Wnt/beta-Catenin Signaling in the Chemoresistance Modulation of Colorectal Cancer. Biomed. Res. Int. 2020, 2020, 9390878. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.Z.; Mei, S.W.; Wang, Z.J.; Chen, J.N.; Shen, H.Y.; Zhao, F.Q.; Li, J.; Xiao, T.X.; Liu, Q. Development and Validation of a Nomogram and a Comprehensive Prognostic Analysis of an LncRNA-Associated Competitive Endogenous RNA Network Based on Immune-Related Genes for Locally Advanced Rectal Cancer With Neoadjuvant Therapy. Front. Oncol. 2021, 11, 697948. [Google Scholar] [CrossRef]
- Zhou, Y.; He, A.; Zhang, L.; Yi, G. MiR-744 mediates the Oxaliplatin chemoresistance in colorectal cancer through inhibiting BIN1. Neoplasma 2020, 67, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Toden, S.; Kunitoshi, S.; Cardenas, J.; Gu, J.; Hutchins, E.; Van Keuren-Jensen, K.; Uetake, H.; Toiyama, Y.; Goel, A. Cancer stem cell-associated miRNAs serve as prognostic biomarkers in colorectal cancer. JCI Insight 2019, 4, e125294. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Shao, B.; Liu, Z.; Dang, Q.; Guo, Y.; Chen, C.; Guo, Y.; Chen, Z.; Liu, J.; Hu, S.; et al. LINC01296/miR-141-3p/ZEB1-ZEB2 axis promotes tumor metastasis via enhancing epithelial-mesenchymal transition process. J. Cancer 2021, 12, 2723–2734. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Jing, H.; Zhang, Y.; Suo, J.; Qian, M. MicroRNA-141-3p affected proliferation, chemosensitivity, migration and invasion of colorectal cancer cells by targeting EGFR. Int. J. Biochem. Cell Biol. 2020, 118, 105643. [Google Scholar] [CrossRef]
- Tanaka, S.; Hosokawa, M.; Yonezawa, T.; Hayashi, W.; Ueda, K.; Iwakawa, S. Induction of epithelial-mesenchymal transition and down-regulation of miR-200c and miR-141 in oxaliplatin-resistant colorectal cancer cells. Biol. Pharm. Bull. 2015, 38, 435–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Liu, Z.; Hu, J.; Luo, Z.; Zhang, C.; Wang, L.; Wang, Z. MiR-377-3p suppresses colorectal cancer through negative regulation on Wnt/beta-catenin signaling by targeting XIAP and ZEB2. Pharmacol. Res. 2020, 156, 104774. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Zhang, J.; Chen, S.; Zhu, J.; Wang, X. Long noncoding RNA CRART16 confers 5-FU resistance in colorectal cancer cells by sponging miR-193b-5p. Cancer Cell Int. 2021, 21, 638. [Google Scholar] [CrossRef]
- Azar, M.; Aghazadeh, H.; Mohammed, H.N.; Sara, M.R.S.; Hosseini, A.; Shomali, N.; Tamjidifar, R.; Tarzi, S.; Mansouri, M.; Sarand, S.P.; et al. miR-193a-5p as a promising therapeutic candidate in colorectal cancer by reducing 5-FU and Oxaliplatin chemoresistance by targeting CXCR4. Int. Immunopharmacol. 2021, 92, 107355. [Google Scholar] [CrossRef]
- Yao, H.; Xia, D.; Li, Z.L.; Ren, L.; Wang, M.M.; Chen, W.S.; Hu, Z.C.; Yi, G.P.; Xu, L. MiR-382 functions as tumor suppressor and chemosensitizer in colorectal cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.; Liu, Y.; Li, Q.; Liu, Q.; Liu, Y.; Luo, Y.; Wei, S. EVs delivery of miR-1915-3p improves the chemotherapeutic efficacy of oxaliplatin in colorectal cancer. Cancer Chemother. Pharmacol. 2021, 88, 1021–1031. [Google Scholar] [CrossRef]
- Sun, W.; Li, J.; Zhou, L.; Han, J.; Liu, R.; Zhang, H.; Ning, T.; Gao, Z.; Liu, B.; Chen, X.; et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics 2020, 10, 1981–1996. [Google Scholar] [CrossRef]
- Tsai, H.L.; Tsai, Y.C.; Chen, Y.C.; Huang, C.W.; Chen, P.J.; Li, C.C.; Su, W.C.; Chang, T.K.; Yeh, Y.S.; Yin, T.C.; et al. MicroRNA-148a induces apoptosis and prevents angiogenesis with bevacizumab in colon cancer through direct inhibition of ROCK1/c-Met via HIF-1alpha under hypoxia. Aging 2022, 14, 6668–6688. [Google Scholar] [CrossRef] [PubMed]
- Khoshinani, H.M.; Afshar, S.; Pashaki, A.S.; Mahdavinezhad, A.; Nikzad, S.; Najafi, R.; Amini, R.; Gholami, M.H.; Khoshghadam, A.; Saidijam, M. Involvement of miR-155/FOXO3a and miR-222/PTEN in acquired radioresistance of colorectal cancer cell line. Jpn. J. Radiol. 2017, 35, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Li, P.L.; Zhang, X.; Wang, L.L.; Du, L.T.; Yang, Y.M.; Li, J.; Wang, C.X. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis 2015, 36, 1484–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Jiang, H.; Zhang, F.; Gao, J.; Hou, J. MicroRNA-330 inhibited cell proliferation and enhanced chemosensitivity to 5-fluorouracil in colorectal cancer by directly targeting thymidylate synthase. Oncol. Lett. 2017, 13, 3387–3394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Chen, X.; Xu, M.; Liu, X.; Pan, B.; Qin, J.; Xu, T.; Zeng, K.; Pan, Y.; He, B.; et al. miR-375-3p suppresses tumorigenesis and partially reverses chemoresistance by targeting YAP1 and SP1 in colorectal cancer cells. Aging 2019, 11, 7357–7385. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Ye, M.L.; Zhang, Y.P.; Li, W.J.; Li, M.T.; Wang, H.Z.; Qiu, X.; Xu, Y.; Yin, J.W.; Hu, Q.; et al. MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci. 2020, 111, 1528–1541. [Google Scholar] [CrossRef]
- Deng, X.; Li, D.; Ke, X.; Wang, Q.; Yan, S.; Xue, Y.; Wang, Q.; Zheng, H. Mir-488 alleviates chemoresistance and glycolysis of colorectal cancer by targeting PFKFB3. J. Clin. Lab. Anal. 2021, 35, e23578. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, H.; Tian, Y.; Li, Y.; Li, J.; Zhong, X.; Yuan, X. LncRNA SNHG11 enhances bevacizumab resistance in colorectal cancer by mediating miR-1207-5p/ABCC1 axis. Anticancer Drugs 2022, 33, 575–586. [Google Scholar] [CrossRef]
- Cui, G.; Zhao, H.; Li, L. Long noncoding RNA PRKCQ-AS1 promotes CRC cell proliferation and migration via modulating miR-1287-5p/YBX1 axis. J. Cell Biochem. 2020, 121, 4166–4175. [Google Scholar] [CrossRef]
- Maier, E.; Attenberger, F.; Tiwari, A.; Lettau, K.; Rebholz, S.; Fehrenbacher, B.; Schaller, M.; Gani, C.; Toulany, M. Dual Targeting of Y-Box Binding Protein-1 and Akt Inhibits Proliferation and Enhances the Chemosensitivity of Colorectal Cancer Cells. Cancers 2019, 11, 562. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.M.; Huang, M.Y.; Chen, Y.C.; Chen, P.J.; Su, W.C.; Chang, T.K.; Li, C.C.; Huang, C.W.; Tsai, H.L.; Wang, J.Y. miRNA-148a Enhances the Treatment Response of Patients with Rectal Cancer to Chemoradiation and Promotes Apoptosis by Directly Targeting c-Met. Biomedicines 2021, 9, 1371. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.C.; Chang, T.K.; Su, W.C.; Tsai, H.L.; Wang, J.Y. Narrative review of the influence of diabetes mellitus and hyperglycemia on colorectal cancer risk and oncological outcomes. Transl. Oncol. 2021, 14, 101089. [Google Scholar] [CrossRef]
- Ning, T.; Li, J.; He, Y.; Zhang, H.; Wang, X.; Deng, T.; Liu, R.; Li, H.; Bai, M.; Fan, Q.; et al. Exosomal miR-208b related with oxaliplatin resistance promotes Treg expansion in colorectal cancer. Mol. Ther. 2021, 29, 2723–2736. [Google Scholar] [CrossRef]
- Han, J.; Sun, W.; Liu, R.; Zhou, Z.; Zhang, H.; Chen, X.; Ba, Y. Plasma Exosomal miRNA Expression Profile as Oxaliplatin-Based Chemoresistant Biomarkers in Colorectal Adenocarcinoma. Front. Oncol. 2020, 10, 1495. [Google Scholar] [CrossRef] [PubMed]
- Pesta, M.; Kucera, R.; Topolcan, O.; Karlikova, M.; Houfkova, K.; Polivka, J.; Macanova, T.; Machova, I.; Slouka, D.; Kulda, V. Plasma microRNA Levels Combined with CEA and CA19-9 in the Follow-Up of Colorectal Cancer Patients. Cancers 2019, 11, 864. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhang, J.; Yao, X.; Jiang, C.; Ni, P.; Cheng, L.; Liu, J.; Ni, S.; Chen, Q.; Li, Q.; et al. Bevacizumab-enhanced antitumor effect of 5-fluorouracil via upregulation of thymidine phosphorylase through vascular endothelial growth factor A/vascular endothelial growth factor receptor 2-specificity protein 1 pathway. Cancer Sci. 2018, 109, 3294–3304. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, M.; Kastan, M.B. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015, 66, 129–143. [Google Scholar] [CrossRef] [Green Version]
- Citrin, D.E.; Mitchell, J.B. Altering the response to radiation: Sensitizers and protectors. Semin. Oncol. 2014, 41, 848–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horak, J.; Dolnikova, A.; Cumaogullari, O.; Cumova, A.; Navvabi, N.; Vodickova, L.; Levy, M.; Schneiderova, M.; Liska, V.; Andera, L.; et al. MiR-140 leads to MRE11 downregulation and ameliorates oxaliplatin treatment and therapy response in colorectal cancer patients. Front. Oncol. 2022, 12, 959407. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.M.; Tsai, H.L.; Chen, Y.C.; Huang, C.W.; Li, C.C.; Su, W.C.; Chang, T.K.; Yeh, Y.S.; Chen, P.J.; Huang, M.Y.; et al. Role of non-coding RNAs in radiosensitivity of colorectal cancer: A narrative review. Front. Oncol. 2022, 12, 889658. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Ding, Y.; Yang, H.; Guo, H.; Zhou, X.; Chen, X. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101-3p sponging. Exp. Mol. Pathol. 2020, 115, 104448. [Google Scholar] [CrossRef] [PubMed]
- Afshar, S.; Najafi, R.; Sedighi Pashaki, A.; Sharifi, M.; Nikzad, S.; Gholami, M.H.; Khoshghadam, A.; Amini, R.; Karimi, J.; Saidijam, M. MiR-185 enhances radiosensitivity of colorectal cancer cells by targeting IGF1R and IGF2. Biomed. Pharmacother. 2018, 106, 763–769. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, J.; Zhou, Z.; He, Z. miR-195 enhances the radiosensitivity of colorectal cancer cells by suppressing CARM1. OncoTargets Ther. 2017, 10, 1027–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Li, L.; Fan, K.; Li, Y.; Gao, Y. A Genome-Scale CRISPR Knock-Out Screen Identifies MicroRNA-5197-5p as a Promising Radiosensitive Biomarker in Colorectal Cancer. Front. Oncol. 2021, 11, 696713. [Google Scholar] [CrossRef]
- Bene, A.; Chambers, T.C. p21 functions in a post-mitotic block checkpoint in the apoptotic response to vinblastine. Biochem. Biophys. Res. Commun. 2009, 380, 211–217. [Google Scholar] [CrossRef] [Green Version]
- Gorospe, M.; Wang, X.; Guyton, K.Z.; Holbrook, N.J. Protective role of p21(Waf1/Cip1) against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol. Cell. Biol. 1996, 16, 6654–6660. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Zhang, Y.; Liu, Y.; Zhou, M.; Lu, Y.; Yuan, L.; Zhang, C.; Hong, M.; Wang, S.; Li, X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J. Transl. Med. 2015, 13, 252. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Liu, J.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Ge, H.; Liu, Y. Exosome-mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J. Exp. Clin. Cancer Res. 2020, 39, 65. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Chen, X.; Liu, J.; Gu, H.; Fan, R.; Ge, H. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 2020, 11, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, H.; Zhang, S.; Zhou, H.; Guo, L. Direct Downregulation of B-Cell Translocation Gene 3 by microRNA-93 Is Required for Desensitizing Esophageal Cancer to Radiotherapy. Dig. Dis. Sci. 2017, 62, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Wu, B.; Zhang, X.; Yang, C.; Zhou, C.; Ren, S.; Wang, J.; Yang, Y.; Wang, G. Screening of MicroRNA Related to Irradiation Response and the Regulation Mechanism of miRNA-96-5p in Rectal Cancer Cells. Front. Oncol. 2021, 11, 699475. [Google Scholar] [CrossRef]
- Rubio, J.; Cristobal, I.; Santos, A.; Carames, C.; Luque, M.; Sanz-Alvarez, M.; Zazo, S.; Madoz-Gurpide, J.; Rojo, F.; Garcia-Foncillas, J. Low MicroRNA-19b Expression Shows a Promising Clinical Impact in Locally Advanced Rectal Cancer. Cancers 2021, 13, 1456. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Yin, Y.F.; Jin, H.G.; Liu, H.R.; Tian, W.C. Exosomal microRNA-19b targets FBXW7 to promote colorectal cancer stem cell stemness and induce resistance to radiotherapy. Kaohsiung J. Med. Sci. 2022, 38, 108–119. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, E.; Fassan, M.; Maretto, I.; Pucciarelli, S.; Zanon, C.; Digito, M.; Rugge, M.; Nitti, D.; Agostini, M. Serum miR-125b is a non-invasive predictive biomarker of the pre-operative chemoradiotherapy responsiveness in patients with rectal adenocarcinoma. Oncotarget 2016, 7, 28647–28657. [Google Scholar] [CrossRef] [PubMed]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.L.; He, G.Y.; Lan, X.L.; Zeng, Z.C.; Guan, J.; Ding, Y.; Qian, X.L.; Liao, W.T.; Ding, Y.Q.; Liang, L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis 2018, 7, 16. [Google Scholar] [CrossRef] [Green Version]
- Salim, H.; Akbar, N.S.; Zong, D.; Vaculova, A.H.; Lewensohn, R.; Moshfegh, A.; Viktorsson, K.; Zhivotovsky, B. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br. J. Cancer 2012, 107, 1361–1373. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Sun, Y.; Yang, Y.; Chen, Y.; Liu, H. Circ_0067835 Knockdown Enhances the Radiosensitivity of Colorectal Cancer by miR-296-5p/IGF1R Axis. OncoTargets Ther. 2021, 14, 491–502. [Google Scholar] [CrossRef]
- Li, L.; Jiang, Z.; Zou, X.; Hao, T. Exosomal circ_IFT80 Enhances Tumorigenesis and Suppresses Radiosensitivity in Colorectal Cancer by Regulating miR-296-5p/MSI1 Axis. Cancer Manag. Res. 2021, 13, 1929–1941. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.B.; Sheng, X.; Zhang, N.; Yang, M.W.; Wang, F. Role of microRNAs in the resistance of colorectal cancer to chemoradiotherapy. Mol. Clin. Oncol. 2018, 8, 523–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef] [PubMed]
- Kelnar, K.; Bader, A.G. A qRT-PCR Method for Determining the Biodistribution Profile of a miR-34a Mimic. Methods Mol. Biol. 2015, 1317, 125–133. [Google Scholar] [CrossRef]

| MiRs Used in the Diagnosis of Primary CRC | Sample Source | Ref. | |
|---|---|---|---|
| miR-20a | MiR-20a was upregulated in patients with CRC (relative to controls) and may be a valid biomarker for CRC detection but may not be a strong prognostic indicator. | Feces, serum, and tumor tissue | [62] |
| miR-21 | The high expression of miR-21 was significantly correlated with advanced clinical stage and poor cell differentiation. | Tumor tissue | [64] |
| miR-29a, miR-223, miR-224 | The expression levels of miR-29a, miR-223, and miR-224 from patients with CRC were significantly lower than those from health volunteers. | Feces | [61] |
| miR-106a, miR-125b | MiR-106a and miR-125b were associated with the pathogenesis and invasion of CRC and may be used as significant prognostic markers of early-stage CRC. | Tumor | [63] |
| miR-129-1-3p, miR-566 | A urinary biomarker panel combining miR-129-1-3p and miR-566 could accurately detect stage 0/I CRC. | Urinary samples | [48] |
| MiRs Used in the Prediction of Early Relapse of CRC | Sample Source | Ref. | |
| miR-21 | Lower serum miR-21 expression was associated with higher local recurrence (p = 0.025) and mortality (p = 0.029). | Serum | [65] |
| miR-29c | miR-29c expression in the early relapse group was significantly lower than that in the non–early relapse group. | Tumor tissue | [66] |
| miR-93 | The miR-93 expression levels of the early relapse group were significantly lower than those of the non–early relapse group. The in vitro and in vivo effects of miR-93 overexpression were inhibited by CRC proliferation and migration, and miR-93 decreased CRC recurrence. | Tumor tissue | [60] |
| miR-148a | miR-148a expression levels in the early relapse group were significantly lower than those in the non–early relapse group. MiR-148a inhibits VEGF secretion by indirectly targeting hypoxia-inducible factor 1 subunit alpha (HIF-1α). | Tumor tissue and serum | [67,68] |
| Lower miR-148a expression was positively associated with advanced TNM stage, poor tumor differentiation, lymph node metastasis, and distant metastasis. | Tumor tissue | [69] | |
| MiRs Used in the Diagnosis of mCRC | Sample Source | Ref. | |
| miR-17/92a-1, miR-106a/363, miR-106b/93/25, miR-183/96/182 clusters | The miR-17/92a-1, miR-106a/363, miR-106b/93/25, and miR-183/96/182 clusters were strongly associated with metastasis and poor patient survival. | Tumor tissue, blood, and feces | [70] |
| miR-20b, miR-29b, miR-155 | A multivariate analysis of patients with mCRC receiving bevacizumab-based treatment revealed that circulating expression levels of miR-20b, miR-29b, and miR-155 were significantly associated with progression-free survival (p < 0.05) and overall survival (p < 0.05). | Serum | [71] |
| miR-96/miR-99b | Plasma miR-96/miR-99b expression levels may serve as a promising biomarker for the early detection of mCRC. | Plasma | [72] |
| miR-210-3p, miR-191-5p, miR-141-3p, miR-1307-5p, miR-155-5p | Five miRNAs—miR-210-3p, miR-191-5p, mir-141-3p, miR-1307-5p, and miR-155-5p—were determined to be upregulated at multiple metastatic sites according to an analysis of new and previously published next-generation sequencing data sets of samples of primary CRC and mCRC (liver, lung, and peritoneal metastases) and tumor-adjacent tissues. | Tumor tissue | [73] |
| miR-762 | The circulating miR-762 levels of patients with CRC with distant metastasis were higher than those of patients with CRC without distant metastasis. | Serum | [74] |
| Family | miRNAs | Verified Targets in CRC or Other Cancers | Sample | Target for miRNA | Ref. |
|---|---|---|---|---|---|
| Let-7 | Let-7a/b/c/d /e/f/g/i, miR-98 | Upregulation of let-7f-5p promotes chemoresistance in CRC by increasing the expression levels of the antiapoptotic proteins B-cell lymphoma 2 (Bcl-2) and B-cell lymphoma extra-large (Bcl-xL) and decreasing the activity of caspase-3 and caspase-9 in CRC cells. | Tumor tissue | p53, p53-inducible nuclear protein 1, p53-inducible nuclear protein 2 and caspase-3. | [84,85] |
| miR-27 | miR-27a, miR-27b | MiR-27a-overexpressing hampered AMPK, enhanced mTOR signaling, unrestricted cell growth, and enhanced chemoresistance. | Tumor tissue | AMPK | [88] |
| miR-96 | miR-96 | Inhibition of miR-96 enhances the sensitivity of CRC cells to oxaliplatin. | Serum | TPM1 | [86] |
| miR-103 | miR-103a/b, miR-107 | MiR-107 induces chemoresistance in CRC cells. | Tumor tissue | CAB39-AMPK-mTOR pathway | [89] |
| MiR-103 and miR-107 enhance chemoresistance in CRC cells by promoting cell stemness. | Wnt/β-catenin signaling | [90] | |||
| miR-744 | miR-744 | The expression levels of miR-744 were significantly elevated in CRC tissues from patients who exhibited preoperative oxaliplatin chemoresistance. MiR-744 may positively mediate oxaliplatin chemoresistance. | Tumor tissue | BIN1 | [92] |
| miR-1246 | miR-1246 | MiR-1246 was overexpressed in CD44v6+ cells and associated with poor overall survival and disease-free survival in patients with CRC. CD44v6+ cells exhibited elevated resistance to chemotherapeutic drugs and significantly higher tumor initiation capacity. | Tumor tissue | DENN/MADD Domain Containing 2D (DENND2D) | [93] |
| Family | miRNAs | Verified Targets in CRC or Other Cancers | Sample Source | Target for miRNA | Ref. |
|---|---|---|---|---|---|
| miR-8 | miR-8, miR-141, miR-200a/b/c, miR-429 | MiR-141-3p enhanced the cetuximab sensitivity of CRC cells | Tumor tissue | ZEB1-ZEB2 | [94,95] |
| Expression of miR-200c and miR-141 was downregulated in oxaliplatin-resistant CRC cell lines. | EGFR | [96] | |||
| miR-27 | miR-27a, miR-27b-3p | MiR-27b-3p sensitizes CRC cells to oxaliplatin in vitro and in vivo, and miR-27b-3p expression was positively correlated with disease-free survival time in patients with CRC. | Tumor tissue | ATG10 | [102] |
| miR-29 | miR-29a, miR-29b, miR-29c | Circulating miR-20b, miR-29b, and miR-155 expression levels were significantly associated with progression-free survival (p < 0.05) and overall survival (p < 0.05). | Serum | No data | [71] |
| miR-34 | miR-34a/b/c/d | MiR-34a enhanced chemosensitivity to 5-FU. | Serum | E2F3; SIRT1. | [87] |
| miR148 | miR-148a/b, miR-152 | MiR-148a suppressed the expression of stem cell markers and increased chemosensitivity, cell invasion, and cell migration. | Tumor tissue | WNT10b and beta-catenin signaling pathway. | [69] |
| MiR-148a decreased angiogenesis and increased CRC cell apoptosis by downregulating HIF-1α/VEGF and Mcl-1, and serum miR-148a levels have prognostic or predictive value in patients with mCRC receiving bevacizumab. | [103] | ||||
| miR-154 | miR-154, miR-323a, miR-369-3p, miR-377, miR-381, miR-382, miR-409, miR-410 | MiR-377-3p expression levels in CRC samples (especially those from patients with stage III/IV CRC) were significantly lower than those in normal mucosa tissues. Overexpression of miR-377-3p enhanced the chemosensitivity of CRC cells by inhibiting Wnt/beta-catenin signaling by directly targeting ZEB2 and XIAP, which are positive regulators of Wnt/β-catenin signaling. | Tumor tissue | ZEB2 and XIAP | [97] |
| MiR-382 functions as a tumor suppressor and chemosensitizer in CRC. | [100] | ||||
| miR-155 | miR-155 | Circulating expression levels of miR-20b, miR-29b, and miR-155 were significantly associated with progression-free survival (p < 0.05) and overall survival (p < 0.05). | Serum | No data | [71] |
| MiR-155 induced radioresistance by targeting FOXO3a. | FOXO3a. | [104] | |||
| miR-193 | miR-193a/b | MiR-193b-5p enhanced chemosensitivity to 5-FU. | Tumor tissue | HMGA2/MAPK pathway | [98] |
| CRC tissues and adjacent noncancerous tissues were obtained from 67 patients who had undergone surgery. Upregulation of miR-193-5p, particularly in combination with 5-FU and oxaliplatin, reduced the expression levels of CXCR4. A miR-193a-5p mimic suppressed CXCR4-induced CRC cell proliferation. | CXCR4. | [99] | |||
| miR-218 | miR-218-1/2 | MiR-218 enhanced 5-FU cytotoxicity by suppressing thymidylate synthase and MiR-218 promoted apoptosis, inhibited cell proliferation, and caused cell cycle arrest | Tumor tissue | thymidylate synthase; BIRC5 | [105] |
| miR-330 | miR-330 | MiR-330 inhibited CRC cell proliferation and enhanced CRC cell chemosensitivity to 5-FU | Tumor tissue | Hexokinase 2Thymidylate synthase | [30] [106] |
| miR-375 | miR-375-3p | MiR-375 enhanced CRC cell chemosensitivity to 5-FU by directly targeting YAP1 and SP1. | Tumor tissue | YAP1 and SP1 | [107] |
| MiR-375 enhanced CRC cell chemosensitivity to 5-FU by targeting thymidylate synthase. | Tumor tissue | thymidylate synthase | [108] | ||
| miR-488 | miR-488 | MiR-488 mimics transfected into CRC cell lines induced decreases in glucose uptake and increases in oxaliplatin/5-FU chemosensistivity. | Serum | PFKFB3 | [109] |
| miR-1207 | miR-1207-5p | Upregulation of miR-1207-5p inhibited bevacizumab resistance in CRC cells. | Tumor tissue | ABCC1. | [110] |
| miR-1287 | miR-1287-5p | Lower miR-1287-5p expression levels upregulate the mRNA expression of Y-box binding protein 1 (YBX1) and protein levels of YBX1, thereby inducing CRC cell proliferation and migration. | Tumor tissue | YBX1 | [111] |
| The multifunctional YBX1 is overexpressed and phosphorylated in CRC and is associated with cetuximab resistance. | [112] | ||||
| miR-1915 | miR-1915 | Exosomal delivery of miR-1915-3p can improve the chemosensitivity of oxaliplatin by suppressing the epithelial–mesenchymal transition. | Plasma | PFKFB3 and USP2 | [101] |
| Family | MiRs | Verified Targets in CRC or Other Cancers | Sample Source | Target for miRNA | Ref. |
|---|---|---|---|---|---|
| miR-10 | miR-10a/b, miR-99a/b, miR-100, miR-125a/b-1/b-2 | MiR-125b was highly expressed both in tissues and serum obtained from nonresponders to CRT. Circulating miR-125b levels exhibited greater discriminatory power for treatment responses than did serum CEA levels. | Serum | No data | [137] |
| miR-155 | miR-155 | Circulating miR-20b, miR-29b, and miR-155 levels were significantly associated with progression-free survival (p < 0.05) and overall survival (p < 0.05). MiR-155 induced radiation resistance. | Serum | No data FOXO3a | [71] [104] |
| miR-214 | miR-214 | MiR-214 promoted radiosensitivity by inhibiting ATG12-mediated autophagy in CRC. | Tumor tissue | ATG12 | [139] |
| miR-296 | miR-296-5p | MiR-296-5p enhanced the radiosensitivity. | Serum | IGF1R; MSI1 | [141] [142] |
| Anti-OncomiRs That Enhance Chemoradiosensitivity | |||||
|---|---|---|---|---|---|
| Family | MiRs | Verified Targets in CRC or Other Cancers | Sample Source | Target for miRNA | Ref. |
| miR-148 | miR-148a/b, miR-152 | MiR-148a enhances the chemoradiosensitivity of patients with rectal cancer. | Tumor tissue | c-Met | [113] |
| miR-34 | miR-34a/b/c/d | MiR-34a attenuates the chemoresistance of colon cancer to 5-FU by inhibiting E2F3 and SIRT1. The miR-34a mimic MRX34 is the first synthetic miRNA to undergo clinical trials. | Serum | E2F3; SIRT1 | [87,143] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, I.-P.; Yip, K.-L.; Chang, Y.-T.; Chen, Y.-C.; Huang, C.-W.; Tsai, H.-L.; Yeh, Y.-S.; Wang, J.-Y. MicroRNAs as Predictive Biomarkers in Patients with Colorectal Cancer Receiving Chemotherapy or Chemoradiotherapy: A Narrative Literature Review. Cancers 2023, 15, 1358. https://doi.org/10.3390/cancers15051358
Yang I-P, Yip K-L, Chang Y-T, Chen Y-C, Huang C-W, Tsai H-L, Yeh Y-S, Wang J-Y. MicroRNAs as Predictive Biomarkers in Patients with Colorectal Cancer Receiving Chemotherapy or Chemoradiotherapy: A Narrative Literature Review. Cancers. 2023; 15(5):1358. https://doi.org/10.3390/cancers15051358
Chicago/Turabian StyleYang, I-Ping, Kwan-Ling Yip, Yu-Tang Chang, Yen-Cheng Chen, Ching-Wen Huang, Hsiang-Lin Tsai, Yung-Sung Yeh, and Jaw-Yuan Wang. 2023. "MicroRNAs as Predictive Biomarkers in Patients with Colorectal Cancer Receiving Chemotherapy or Chemoradiotherapy: A Narrative Literature Review" Cancers 15, no. 5: 1358. https://doi.org/10.3390/cancers15051358
APA StyleYang, I. -P., Yip, K. -L., Chang, Y. -T., Chen, Y. -C., Huang, C. -W., Tsai, H. -L., Yeh, Y. -S., & Wang, J. -Y. (2023). MicroRNAs as Predictive Biomarkers in Patients with Colorectal Cancer Receiving Chemotherapy or Chemoradiotherapy: A Narrative Literature Review. Cancers, 15(5), 1358. https://doi.org/10.3390/cancers15051358