High-Multiplex Aptamer-Based Serum Proteomics to Identify Candidate Serum Biomarkers of Oral Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
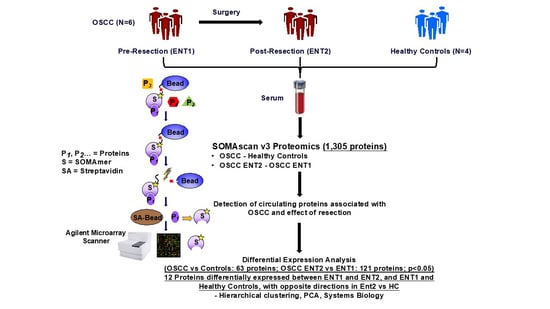
2. Materials and Methods
2.1. Sample Collection
2.2. SOMAScan Analysis
2.3. Quantification of Biomarker Candidates
2.4. Bioinfornatics and Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagata, M.; Kurita, H.; Uematsu, K.; Ogawa, S.; Takahashi, K.; Hoshina, H.; Takagi, R. Diagnostic value of cyclin-dependent kinase/cyclin-dependent kinase inhibitor expression ratios as biomarkers of locoregional and hematogenous dissemination risks in oral squamous cell carcinoma. Mol. Clin. Oncol. 2015, 3, 1007–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, P.Y.; Hsu, N.C.; Lin, S.H.; Hou, M.F.; Yeh, K.T. High nuclear protein kinase CbetaII expression is a marker of disease recurrence in oral squamous cell carcinoma. Anticancer Res. 2012, 32, 3987–3991. [Google Scholar] [PubMed]
- Seki, S.; Fujiwara, M.; Matsuura, M.; Fujita, S.; Ikeda, H.; Asahina, I.; Ikeda, T. Prediction of outcome of patients with oral squamous cell carcinoma using vascular invasion and the strongly positive expression of vascular endothelial growth factors. Oral Oncol. 2011, 47, 588–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.-Y.; Hsi, E.; Huang, Y.-C.; Hsu, N.C.-H.; Chu, P.-Y.; Chai, C.-Y. High LC3 expression correlates with poor survival in patients with oral squamous cell carcinoma. Hum. Pathol. 2013, 44, 2558–2562. [Google Scholar] [CrossRef]
- Matos, L.L.; Dedivitis, R.A.; Kulcsar, M.A.V.; de Mello, E.S.; Alves, V.A.F.; Cernea, C.R. External validation of the AJCC Cancer Staging Manual, 8th edition, in an independent cohort of oral cancer patients. Oral Oncol. 2017, 71, 47–53. [Google Scholar] [CrossRef]
- Fu, T.-Y.; Hsieh, I.-C.; Cheng, J.-T.; Tsai, M.-H.; Hou, Y.-Y.; Lee, J.-H.; Liou, H.-H.; Huang, S.-F.; Chen, H.-C.; Yen, L.-M.; et al. Association of OCT4, SOX2, and NANOG expression with oral squamous cell carcinoma progression. J. Oral Pathol. Med. 2016, 45, 89–95. [Google Scholar] [CrossRef]
- Oliveira-Costa, J.P.; de Carvalho, A.F.; da Silveira, G.G.; Amaya, P.; Wu, Y.; Park, K.J.; Gigliola, M.P.; Lustberg, M.; Buim, M.E.; Ferreira, E.N.; et al. Gene expression patterns through oral squamous cell carcinoma development: PD-L1 expression in primary tumor and circulating tumor cells. Oncotarget 2015, 6, 20902–20920. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Liu, Y.; Johnson, J.J.; Stack, M.S. Urinary-type plasminogen activator receptor (uPAR) modulates oral cancer cell behavior with alteration in p130cas. Mol. Cell. Biochem. 2011, 357, 151–161. [Google Scholar] [CrossRef]
- Huber, G.F.; Albinger-Hegyi, A.; Soltermann, A.; Roessle, M.; Graf, N.; Haerle, S.K.; Holzmann, D.; Moch, H.; Hegyi, I. Expression patterns of Bmi-1 and p16 significantly correlate with overall, disease-specific, and recurrence-free survival in oropha-ryngeal squamous cell carcinoma. Cancer 2011, 117, 4659–4670. [Google Scholar] [CrossRef]
- Blatt, S.; Krüger, M.; Ziebart, T.; Sagheb, K.; Schiegnitz, E.; Goetze, E.; Al-Nawas, B.; Pabst, A.M. Biomarkers in diagnosis and therapy of oral squamous cell carcinoma: A review of the literature. J. Cranio-Maxillofac. Surg. 2017, 45, 722–730. [Google Scholar] [CrossRef]
- Sartini, D.; Pozzi, V.; Renzi, E.; Morganti, S.; Rocchetti, R.; Rubini, C.; Santarelli, A.; Muzio, L.L.; Emanuelli, M. Analysis of tissue and salivary nicotinamide N-methyltransferase in oral squamous cell carcinoma: Basis for the development of a noninvasive diagnostic test for early-stage disease. Biol. Chem. 2012, 393, 505–511. [Google Scholar] [CrossRef]
- Jiang, T.; Liu, G.; Wang, L.; Liu, H. Elevated Serum Gas6 Is a Novel Prognostic Biomarker in Patients with Oral Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0133940. [Google Scholar] [CrossRef]
- Gallo, C.; Ciavarella, M.; Santarelli, A.; Ranieri, E.; Colella, G.; Muzio, L.L.; Russo, L.L. Potential Salivary Proteomic Markers of Oral Squamous Cell Carcinoma. Cancer Genom. Proteom. 2015, 13, 55–61. [Google Scholar]
- Soltys, S.G.; Le, Q.T.; Shi, G.; Tibshirani, R.; Giaccia, A.J.; Koong, A.C. The use of plasma surface-enhanced laser desorp-tion/ionization time-of-flight mass spectrometry proteomic patterns for detection of head and neck squamous cell cancers. Clin. Cancer Res. 2004, 10, 4806–4812. [Google Scholar] [CrossRef] [Green Version]
- Wadsworth, J.T.; Somers, K.D.; Stack, J.B.C.; Cazares, L.; Malik, G.; Adam, B.-L.; Wright, J.G.L.; Semmes, O.J. Identification of Patients With Head and Neck Cancer Using Serum Protein Profiles. Arch. Otolaryngol. Neck Surg. 2004, 130, 98–104. [Google Scholar] [CrossRef] [Green Version]
- Gourin, C.G.; Zhi, W.; Adam, B.-L. Proteomic identification of serum biomarkers for head and neck cancer surveillance. Laryngoscope 2009, 119, 1291–1302. [Google Scholar] [CrossRef]
- Palstrøm, N.B.; Matthiesen, R.; Rasmussen, L.M.; Beck, H.C. Recent Developments in Clinical Plasma Proteomics—Applied to Cardiovascular Research. Biomedicines 2022, 10, 162. [Google Scholar] [CrossRef]
- Schuppan, D.; Myneni, S.; Surabattula, R. Liquid biomarkers for fibrotic NASH—Progress in a complex field. J. Hepatol. 2022, 76, 5–7. [Google Scholar] [CrossRef]
- Gold, L.; Walker, J.J.; Wilcox, S.K.; Williams, S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. New Biotechnol. 2012, 29, 543–549. [Google Scholar] [CrossRef]
- Dayon, L.; Cominetti, O.; Affolter, M. Proteomics of human biological fluids for biomarker discoveries: Technical advances and recent applications. Expert Rev. Proteom. 2022, 19, 131–151. [Google Scholar] [CrossRef]
- Shubin, A.V.; Kollar, B.; Dillon, S.T.; Pomahac, B.; Libermann, T.A.; Riella, L.V. Blood proteome profiling using aptamer-based technology for rejection biomarker discovery in transplantation. Sci. Data 2019, 6, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, Z.; Pan, X.; Parlayan, C.; Ojima, H.; Kondo, T. Proteomic study of hepatocellular carcinoma using a novel modified ap-tamer-based array (SOMAscan) platform. Biochim. Biophys. Acta 2017, 1865, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Olaleye, O.; Govorukhina, N.; van de Merbel, N.C.; Bischoff, R. Non-Antibody-Based Binders for the Enrichment of Proteins for Analysis by Mass Spectrometry. Biomolecules 2021, 11, 1791. [Google Scholar] [CrossRef] [PubMed]
- Vasunilashorn, S.M.; Dillon, S.T.; Chan, N.Y.; Fong, T.G.; Joseph, M.; Tripp, B.; Xie, Z.; Ngo, L.H.; Lee, C.G.; Elias, J.A.; et al. Proteome-Wide Analysis Using SOMAscan Identifies and Validates Chitinase-3-like Protein 1 as a Risk and Disease Marker of Delirium among Older Adults Undergoing Major Elective Surgery. J. Gerontol. A Biol. Sci. Med. Sci. 2021, 77, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Ostroff, R.; Hinterberg, M.A.; Coresh, J.; Ballantyne, C.M.; Matsushita, K.; Mueller, C.E.; Walter, J.; Jonasson, C.; Holman, R.R.; et al. A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk. Sci. Transl. Med. 2022, 14, eabj9625. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [Green Version]
- Webber, J.; Stone, T.C.; Katilius, E.; Smith, B.C.; Gordon, B.; Mason, M.D.; Tabi, Z.; Brewis, I.A.; Clayton, A. Proteomics Analysis of Cancer Exosomes Using a Novel Modified Aptamer-based Array (SOMAscanTM) Platform. Mol. Cell. Proteom. 2014, 13, 1050–1064. [Google Scholar] [CrossRef] [Green Version]
- Mehan, M.R.; Williams, S.A.; Siegfried, J.M.; Bigbee, W.L.; Weissfeld, J.L.; Wilson, D.O.; Pass, H.I.; Rom, W.N.; Muley, T.; Meister, M.; et al. Validation of a blood protein signature for non-small cell lung cancer. Clin. Proteom. 2014, 11, 32. [Google Scholar] [CrossRef] [Green Version]
- Hathout, Y.; Brody, E.; Clemens, P.R.; Cripe, L.; DeLisle, R.K.; Furlong, P.; Gordish-Dressman, H.; Hache, L.; Henricson, E.; Hoffman, E.P.; et al. Large-scale serum protein biomarker discovery in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 2015, 112, 7153–7158. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Carnielli, C.M.; Macedo, C.C.S.; De Rossi, T.; Granato, D.C.; Rivera, C.; Domingues, R.R.; Pauletti, B.A.; Yokoo, S.; Heberle, H.; Busso-Lopes, A.F.; et al. Combining discovery and targeted proteomics reveals a prognostic signature in oral cancer. Nat. Commun. 2018, 9, 3598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.W.; Chang, K.P.; Yen, W.C.; Liu, H.P.; Chan, X.Y.; Liu, C.R.; Hung, C.M.; Wu, C.C. Identification of salivary autoanti-bodies as biomarkers of oral cancer with immunoglobulin A enrichment combined with affinity mass spectrometry. Proteomics 2023, e2200321. [Google Scholar] [CrossRef]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Thorsen, S.B.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-plex PEA immuno-assay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higgins, S.J.; De Ceunynck, K.; Kellum, J.A.; Chen, X.; Gu, X.; Chaudhry, S.A.; Schulman, S.; Libermann, T.A.; Lu, S.; Shapiro, N.I.; et al. Tie2 protects the vasculature against thrombus formation in systemic in-flammation. J. Clin. Investig. 2018, 128, 1471–1484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Chen, X.; Fu, X.; Li, Z.; Huang, Y.; Liang, C. Advances in Aptamer-Based Biomarker Discovery. Front. Cell Dev. Biol. 2021, 9, 659760. [Google Scholar] [CrossRef]
- Lindbohm, J.V.; Mars, N.; Walker, K.A.; Singh-Manoux, A.; Livingston, G.; Brunner, E.J.; Sipilä, P.N.; Saksela, K.; Ferrie, J.E.; Lovering, R.C.; et al. Plasma proteins, cognitive decline, and 20-year risk of dementia in the Whitehall II and Atherosclerosis Risk in Communities studies. Alzheimers Dement. 2022, 18, 612–624. [Google Scholar] [CrossRef]
- Muruve, D.A.; Debiec, H.; Dillon, S.T.; Gu, X.; Plaisier, E.; Can, H.; Otu, H.H.; Libermann, T.A.; Ronco, P. Serum Protein Signa-tures Using Aptamer-Based Proteomics for Minimal Change Disease and Membranous Nephropathy. Kidney Int. Rep. 2022, 7, 1539–1556. [Google Scholar] [CrossRef]
- Sasamoto, N.; Ngo, L.; Vitonis, A.F.; Dillon, S.T.; A Missmer, S.; A Libermann, T.; Terry, K.L. Circulating proteomic profiles associated with endometriosis in adolescents and young adults. Hum. Reprod. 2022, 37, 2042–2053. [Google Scholar] [CrossRef]
- Kobayashi, H.; Looker, H.C.; Satake, E.; D’addio, F.; Wilson, J.M.; Saulnier, P.J.; Dom, Z.I.M.; O’Neil, K.; Ihara, K.; Krolewski, B.; et al. Neuroblastoma suppressor of tumorigenicity 1 is a circulating protein associated with progression to end-stage kidney disease in diabetes. Sci. Transl. Med. 2022, 14, abj210. [Google Scholar] [CrossRef]
- Pietzner, M.; Wheeler, E.; Carrasco-Zanini, J.; Kerrison, N.D.; Oerton, E.; Koprulu, M.; Luan, J.; Hingorani, A.D.; Williams, S.A.; Wareham, N.J.; et al. Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat. Commun. 2021, 12, 6822. [Google Scholar] [CrossRef]
- Graumann, J.; Finkernagel, F.; Reinartz, S.; Stief, T.; Brödje, D.; Renz, H.; Jansen, J.M.; Wagner, U.; Worzfeld, T.; Pogge von Strandmann, E.; et al. Multi-platform Affinity Proteomics Identify Proteins Linked to Metastasis and Immune Suppression in Ovarian Cancer Plasma. Front. Oncol. 2019, 9, 1150. [Google Scholar] [CrossRef] [Green Version]
- Finkernagel, F.; Reinartz, S.; Schuldner, M.; Malz, A.; Jansen, J.M.; Wagner, U.; Worzfeld, T.; Graumann, J.; Von Strandmann, E.P.; Müller, R. Dual-platform affinity proteomics identifies links between the recurrence of ovarian carcinoma and proteins released into the tumor microenvironment. Theranostics 2019, 9, 6601–6617. [Google Scholar] [CrossRef]
- Jung, Y.J.; Oh, I.-J.; Kim, Y.; Jung, J.H.; Seok, M.; Lee, W.; Park, C.K.; Lim, J.-H.; Kim, Y.-C.; Kim, W.-S.; et al. Clinical Validation of a Protein Biomarker Panel for Non-Small Cell Lung Cancer. J. Korean Med. Sci. 2018, 33, e342. [Google Scholar] [CrossRef]
- Thanasupawat, T.; Glogowska, A.; Pascoe, C.; Krishnan, S.N.; Munir, M.; Begum, F.; Beiko, J.; Krcek, J.; Del Bigio, M.R.; Pitz, M.; et al. Slow Off-Rate Modified Aptamer (SOMAmer) Proteomic Analysis of Patient-Derived Malignant Glioma Identifies Distinct Cellular Proteomes. Int. J. Mol. Sci. 2021, 22, 9566. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, N.; Ji, N.; Chen, Z.-S. Proteomics technologies for cancer liquid biopsies. Mol. Cancer 2022, 21, 53. [Google Scholar] [CrossRef]
- Kim, C.H.; Tworoger, S.S.; Stampfer, M.J.; Dillon, S.T.; Gu, X.; Sawyer, S.J.; Chan, A.T.; Libermann, T.A.; Eliassen, A.H. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci. Rep. 2018, 8, 8382. [Google Scholar] [CrossRef] [Green Version]
- Raffield, L.M.; Dang, H.; Pratte, K.A.; Jacobson, S.; Gillenwater, L.A.; Ampleford, E.; Barjaktarevic, I.; Basta, P.; Clish, C.B.; Comellas, A.P.; et al. Comparison of Proteomic Assessment Methods in Multiple Cohort Studies. Proteomics 2020, 20, e1900278. [Google Scholar] [CrossRef]
- Gao, Z.-B.; Duan, Y.-Q.; Zhang, L.; Chen, D.-W.; Ding, P.-T. Expression of matrix metalloproteinase 2 and its tissue inhibitor in oral squamous cell carcinoma. Int. J. Mol. Med. 2005, 16, 599–603. [Google Scholar]
- Wang, S.H.; Chen, Y.L.; Hsiao, J.R.; Tsai, F.Y.; Jiang, S.S.; Lee, A.Y.; Tsai, H.J.; Chen, Y.W. Insulin-like growth factor binding pro-tein 3 promotes radiosensitivity of oral squamous cell carcinoma cells via positive feedback on NF-κB/IL-6/ROS signaling. J. Exp. Clin. Cancer Res. 2021, 40, 95. [Google Scholar] [CrossRef]
- Li, C.; Sun, C.-J.; Fan, J.-C.; Geng, N.; Li, C.-H.; Liao, J.; Mi, K.; Zhu, G.-Q.; Ma, H.; Song, Y.-F.; et al. Angiopoietin-2 expression is correlated with angiogenesis and overall survival in oral squamous cell carcinoma. Med. Oncol. 2013, 30, 571. [Google Scholar] [CrossRef]
- Shpitzer, T.; Bahar, G.; Feinmesser, R.; Nagler, R.M. A comprehensive salivary analysis for oral cancer diagnosis. J. Cancer Res. Clin. Oncol. 2007, 133, 613–617. [Google Scholar] [CrossRef] [PubMed]
- Monea, M.; Pop, A.M. The Use of Salivary Levels of Matrix Metalloproteinases as an Adjuvant Method in the Early Diagnosis of Oral Squamous Cell Carcinoma: A Narrative Literature Review. Curr. Issues Mol. Biol. 2022, 44, 6306–6322. [Google Scholar] [CrossRef] [PubMed]
- Sasahira, T.; Kurihara-Shimomura, M.; Shimomura, H.; Bosserhoff, A.K.; Kirita, T. Identification of oral squamous cell carcinoma markers MUC2 and SPRR1B downstream of TANGO. J. Cancer Res. Clin. Oncol. 2021, 147, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.C.; Huang, M.Y.; Chen, Y.K.; Lan, W.C.; Shieh, T.M.; Shih, Y.H. Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenz[a]anthracene-Induced Oral Cancer with Radiation Therapy—A Syrian Golden Hamster Model. Diagnostics 2021, 12, 65. [Google Scholar] [CrossRef]
- Tomasovic-Loncaric, C.; Fucic, A.; Andabak, A.; Andabak, M.; Ceppi, M.; Bruzzone, M.; Vrdoljak, D.; Vucicevic-Boras, V. An-drogen Receptor as a Biomarker of Oral Squamous Cell Carcinoma Progression Risk. Anticancer Res. 2019, 39, 4285–4289. [Google Scholar] [CrossRef]
- Pérez-Sayáns, M.; Suárez-Peñaranda, J.M.; Pilar, G.D.; Barros-Angueira, F.; Gándara-Rey, J.M.; García-García, A. What real in-fluence does the proto-oncogene c-myc have in OSCC behavior? Oral Oncol. 2011, 47, 688–692. [Google Scholar] [CrossRef]
- Dourado, M.R.; Elseragy, A.; da Costa, B.C.; Téo, F.H.; Guimarães, G.N.; Machado, R.A.; Risteli, M.; Wahbi, W.; Rocha, C.A.G.; Paranaíba, L.M.R.; et al. Stress induced phosphoprotein 1 overexpression controls proliferation, migration and invasion and is associated with poor survival in oral squamous cell carcinoma. Front. Oncol. 2023, 12, 1085917. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara-Shimomura, M.; Shimomura, H.; Kirita, T. SERPINE2 is an oral cancer-promoting factor that induces angiogenesis and lymphangiogenesis. Int. J. Clin. Oncol. 2021, 26, 1831–1839. [Google Scholar] [CrossRef]
- Mohajertehran, F.; Ayatolahi, H.; Khazaeni, K.; Shakeri, M.T.; Mohtasham, N. Overexpression of High-Mobility Motor Box 1 in the Blood and Tissues of Patients with Head and Neck Squamous Cell Carcinoma. Iran J. Otorhinolaryngol. 2018, 30, 261–271. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Okui, T.; Yoneda, T.; Ryumon, S.; Nakamura, T.; Kawai, H.; Kunisada, Y.; Ibaragi, S.; Masui, M.; Ono, K.; et al. High-mobility group box 1 induces bone destruction associated with advanced oral squamous cancer via RAGE and TLR4. Biochem. Biophys. Res. Commun. 2020, 531, 422–430. [Google Scholar] [CrossRef]
- Kroemer, G.; Kepp, O. Radiochemotherapy-induced elevations of plasma HMGB1 levels predict therapeutic responses in cancer patients. Oncoimmunology 2021, 10, 2005859. [Google Scholar] [CrossRef]
- Van Teijlingen, E.; Hundley, V. The importance of pilot studies. Nurs. Stand. 2002, 16, 33–36. [Google Scholar] [CrossRef] [Green Version]







| Patient | Gender | Age | Comorbidity | Tumor Site | Status | Grading | Adjuvant Therapy | ||
|---|---|---|---|---|---|---|---|---|---|
| T | N | M | |||||||
| 1 | Male | 70 | Coronary heart disease | Floor of the mouth | 4 | 0 | 0 | 2 | Irradiation (64 Gray) |
| 2 | Female | 63 | None | Cheek | 2 | 1 | 0 | 2 | Irradiation (64 Gray) |
| 3 | Male | 62 | None | Floor of the mouth | 4 | 0 | 0 | 2 | Irradiation (64 Gray) |
| 4 | Male | 80 | Chronic venous disease | Hard palate | 2 | 0 | 0 | 2 | none |
| 5 | Male | 68 | Arterial hypertension | Floor of the mouth | 4 | 2 | 0 | 3 | Irradiation (64 Gray) |
| 6 | Male | 63 | None | Soft palate | 1 | 0 | 0 | 2 | none |
| 7 | Male | 63 | Arterial hypertension | - | - | - | - | - | - |
| 8 | Male | 75 | Depression | - | - | - | - | - | - |
| 9 | Female | 68 | None | - | - | - | - | - | - |
| 10 | Female | 85 | Arterial hypertension, Coronary heart disease | - | - | - | - | - | - |
| Down-regulated Proteins between OSCC and Healthy Control | Up-regulated Proteins between OSCC and Healthy Control |
|---|---|
| Adenylate kinase isoenzyme 1 | Heterogeneous nuclear ribonucleoprotein K |
| Hemoglobin | Complement C5 |
| Xaa-Pro aminopeptidase 1 | Complement factor I |
| Alcohol dehydrogenase [NADP(+)] | Complement factor H |
| Casein kinase II 2-alpha:2-beta heterotetramer | Complement component 1 Q subcomponent-binding protein, mitochondrial |
| Stress-induced-phosphoprotein 1 | Syntaxin-1A |
| Peroxiredoxin-1 | Myc proto-oncogene protein |
| Interleukin-3 receptor subunit alpha | Serine protease 27 |
| Interstitial collagenase | Mammaglobin-B |
| Desert hedgehog protein N-product | Coagulation Factor X |
| Glucocorticoid receptor | Thymidine kinase, cytosolic |
| Metalloproteinase inhibitor 2 | Dynactin subunit 2 |
| Glia-derived nexin | Lymphocyte antigen 86 |
| Kallikrein-8 | Peroxisomal targeting signal 1 receptor |
| Gelsolin | Legumain |
| Importin subunit alpha-1 | Ubiquitin-conjugating enzyme E2 G2 |
| Membrane frizzled-related protein | C3a anaphylatoxin des Arginine |
| Apoptosis regulator Bcl-2 | Complement C1q subcomponent |
| MAP kinase-activated protein kinase 3 | Cadherin-3 |
| A disintegrin and metalloproteinase with thrombospondin motifs 1 | Parathyroid hormone-related protein |
| Cadherin-6 | |
| Kremen protein 2 | HERV-H LTR-associating protein 2 |
| Semaphorin-3A | |
| Mitogen-activated protein kinase 13 | |
| Complement factor B | |
| Angiostatin | |
| Growth/differentiation factor 9 | |
| Thrombin | |
| Aspartate aminotransferase, cytoplasmic | |
| Granulocyte colony-stimulating factor receptor | |
| Endothelin-converting enzyme 1 | |
| Granulins | |
| Mediator of RNA polymerase II transcription subunit 1 | |
| Lysosomal protective protein | |
| SLAM family member 6 | |
| Protein SET | |
| Interleukin-1 Receptor accessory protein | |
| Hepatocyte growth factor activator | |
| Acidic leucine-rich nuclear phosphoprotein 32 family member B | |
| ATP synthase subunit beta, mitochondrial | |
| E3 SUMO-protein ligase PIAS4 | |
| Serum amyloid A-1 protein |
| Increased in ENT2 in OSCC | Decreased in ENT2 in OSCC |
|---|---|
| Ck-beta-8-1 | Creatine kinase M-type:Creatine kinase B-type heterodimer |
| Osteomodulin | Macrophage migration inhibitory factor |
| Macrophage mannose receptor 1 | Tissue Factor |
| Ephrin type-A receptor 2 | Beta-Ala-His dipeptidase |
| Lymphocyte activation gene 3 protein | SPARC |
| SLIT and NTRK-like protein 5 | Thrombospondin-1 |
| Dual specificity mitogen-activated protein kinase kinase 4 | MAP kinase-activated protein kinase 3 |
| Hepatitis A virus cellular receptor 2 | Complement factor H-related protein 5 |
| Netrin receptor UNC5D | Myosin-binding protein C, slow-type |
| Ephrin-B2 | Membrane frizzled-related protein |
| Advanced glycosylation end product-specific receptor, soluble | Estradiol 17-beta-dehydrogenase 1 |
| alpha-Fetoprotein | |
| Tumor necrosis factor ligand superfamily member 15 | Interstitial collagenase |
| Ephrin-A5 | Tyrosine-protein kinase Tec |
| 72 kDa type IV collagenase | Calcium/calmodulin-dependent protein kinase kinase 1 |
| Tumor necrosis factor receptor superfamily member 1B | Hexokinase-1 |
| Antileukoproteinase | Heterogeneous nuclear ribonucleoprotein A/B |
| Spondin-1 | Basigin |
| Follistatin-related protein 3 | Amyloid beta A4 protein |
| Dickkopf-related protein 3 | C-type lectin domain family 7 member A |
| Agouti-related protein | Erythropoietin receptor |
| Mediator of RNA polymerase II transcription subunit 1 | Thrombopoietin Receptor |
| Semaphorin-6A | Bone morphogenetic protein 10 |
| Ephrin-A4 | Activated Protein C |
| Ciliary neurotrophic factor receptor subunit alpha | Serine/threonine-protein kinase TBK1 |
| Periostin | Angiotensin-converting enzyme 2 |
| Thrombospondin-2 | Relaxin receptor 1 |
| Cytochrome c | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform |
| Glutamate carboxypeptidase 2 | |
| Aggrecan core protein | Cystatin-S |
| Programmed cell death 1 ligand 2 | Mesothelin |
| Low affinity immunoglobulin gamma Fc region receptor III-B | Immunoglobulin superfamily containing leucine-rich repeat protein 2 |
| Biglycan | Serine/threonine-protein kinase PLK1 |
| Kallikrein-8 | 40S ribosomal protein S3a |
| WAP, Kazal, immunoglobulin, Kunitz and NTR domain-containing protein 2 | Protein Wnt-7a |
| Brain natriuretic peptide 32 | |
| Decorin | Eukaryotic translation initiation factor 4E-binding protein 2 |
| CMRF35-like molecule 6 | cAMP-regulated phosphoprotein 19 |
| Coagulation Factor V | 40S ribosomal protein S3 |
| SLAM family member 7 | Killer cell lectin-like receptor subfamily F member 1 |
| Thioredoxin domain-containing protein 12 | Microtubule-associated protein tau |
| Tyrosine-protein phosphatase non-receptor type substrate 1 | Killer cell immunoglobulin-like receptor 3DS1 |
| Histone acetyltransferase type B catalytic subunit | |
| Angiostatin | Gastrin-releasing peptide |
| Dynactin subunit 2 | Interleukin-36 beta |
| Bone morphogenetic protein 6 | G2/mitotic-specific cyclin-B1 |
| Cathepsin B | Protein jagged-2 |
| Layilin | AMP Kinase (alpha1beta1gamma1) |
| MHC class I polypeptide-related sequence B | Proteasome activator complex subunit 3 |
| Pigment epithelium-derived factor | Serum paraoxonase/arylesterase 1 |
| Interleukin-15 receptor subunit alpha | Growth/differentiation factor 8 |
| Sialic acid-binding Ig-like lectin 14 | Olfactomedin-4 |
| Wnt inhibitory factor 1 | Ectonucleoside triphosphate diphosphohydrolase 1 |
| Immunoglobulin A | MAP kinase-activated protein kinase 5 |
| Arylsulfatase A | |
| C-type lectin domain family 4 member M | |
| Tyrosine-protein kinase ABL1 | |
| Lysosome membrane protein 2 | |
| A disintegrin and metalloproteinase with thrombospondin motifs 4 | |
| Methyl-CpG-binding domain protein 4 | |
| Cardiotrophin-1 | |
| Proliferating cell nuclear antigen | |
| Baculoviral IAP repeat-containing protein 7 Isoform beta | |
| Neuregulin-4 | |
| Transforming growth factor beta-1 | |
| Interleukin-20 | |
| Fibroblast growth factor receptor 2 | |
| TATA-box-binding protein | |
| Interleukin-16 | |
| Interleukin-17A | |
| Galectin-4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blatt, S.; Kämmerer, P.W.; Krüger, M.; Surabattula, R.; Thiem, D.G.E.; Dillon, S.T.; Al-Nawas, B.; Libermann, T.A.; Schuppan, D. High-Multiplex Aptamer-Based Serum Proteomics to Identify Candidate Serum Biomarkers of Oral Squamous Cell Carcinoma. Cancers 2023, 15, 2071. https://doi.org/10.3390/cancers15072071
Blatt S, Kämmerer PW, Krüger M, Surabattula R, Thiem DGE, Dillon ST, Al-Nawas B, Libermann TA, Schuppan D. High-Multiplex Aptamer-Based Serum Proteomics to Identify Candidate Serum Biomarkers of Oral Squamous Cell Carcinoma. Cancers. 2023; 15(7):2071. https://doi.org/10.3390/cancers15072071
Chicago/Turabian StyleBlatt, Sebastian, Peer W. Kämmerer, Maximilian Krüger, Rambabu Surabattula, Daniel G. E. Thiem, Simon T. Dillon, Bilal Al-Nawas, Towia A. Libermann, and Detlef Schuppan. 2023. "High-Multiplex Aptamer-Based Serum Proteomics to Identify Candidate Serum Biomarkers of Oral Squamous Cell Carcinoma" Cancers 15, no. 7: 2071. https://doi.org/10.3390/cancers15072071
APA StyleBlatt, S., Kämmerer, P. W., Krüger, M., Surabattula, R., Thiem, D. G. E., Dillon, S. T., Al-Nawas, B., Libermann, T. A., & Schuppan, D. (2023). High-Multiplex Aptamer-Based Serum Proteomics to Identify Candidate Serum Biomarkers of Oral Squamous Cell Carcinoma. Cancers, 15(7), 2071. https://doi.org/10.3390/cancers15072071