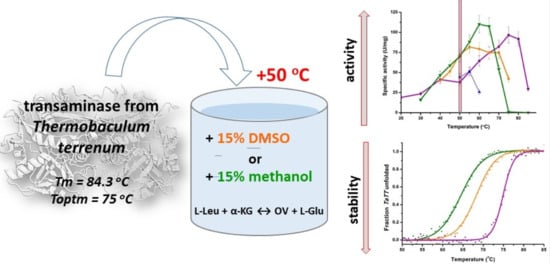
Counterbalance of Stability and Activity Observed for Thermostable Transaminase from Thermobaculum terrenum in the Presence of Organic Solvents
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Enzyme Production and Activity Assays
3.2. Analysis of TaTT Stability
3.3. Analysis of Hydrogen Bonds
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Dimer of | ||||
|---|---|---|---|---|
| TaTT | TA from T. uzoniensis | TA from B. pseudomalle | TA from E. coli | |
| Number of residues per subunit | 316 | 295 | 307 | 309 |
Hydrogen bonds
| 647/648 1.025 293/296 177/179 430/434 77/68 140/146 18/24 | 578 0.98 302 135 416 36 126 14 | 617 1.005 296 151 432 47 138 23 | 554 0.896 287 141 400 49 105 12 |
| PDB ID: | 6GKR | 5CE8 | 3U0G | 1I1K |
References
- Slabu, I.; Galman, J.L.; Lloyd, R.C.; Turner, N.J. Discovery, engineering, and synthetic application of transaminase biocatalysts. ACS Catal. 2017, 7, 8263–8284. [Google Scholar] [CrossRef]
- Savile, C.K.; Janey, J.M.; Mundorff, E.C.; Moore, J.C.; Tam, S.; Jarvis, W.R.; Colbeck, J.C.; Krebber, A.; Fleitz, F.J.; Brands, J.; et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 2010, 329, 305–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doukyu, N.; Ogino, H. Organic solvent-tolerant enzymes. Biochem. Eng. J. 2010, 48, 270–282. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Petrova, T.E.; Popinako, A.V.; Antonov, M.Y.; Stekhanova, T.N.; Popov, V.O. Intramolecular hydrogen bonding in the polyextremophilic short-chain dehydrogenase from the archaeon Thermococcus sibiricus and its close structural homologs. Biochimie 2015, 118, 82–89. [Google Scholar] [CrossRef]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef] [Green Version]
- Stepankova, V.; Bidmanova, S.; Koudelakova, T.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Strategies for stabilization of enzymes in organic solvents. ACS Catal. 2013, 3, 2823–2836. [Google Scholar] [CrossRef]
- Feller, G. Protein stability and enzyme activity at extreme biological temperatures. J. Phys. Condens. Matter 2010, 22, 323101. [Google Scholar] [CrossRef]
- Kumar, S.; Tsai, C.-J.; Ma, B.; Nussinov, R. Contribution of salt bridges toward protein thermostability. J. Biomol. Struct. Dyn. 2000, 17, 79–85. [Google Scholar] [CrossRef]
- Berezovsky, I.N.; Shakhnovich, E.I. Physics and evolution of thermophilic adaptation. Proc. Natl. Acad. Sci. USA 2005, 102, 12742–12747. [Google Scholar] [CrossRef] [Green Version]
- Matsui, I.; Harata, K. Implication for buried polar contacts and ion pairs in hyperthermostable enzymes. FEBS J. 2007, 274, 4012–4022. [Google Scholar] [CrossRef] [Green Version]
- Elcock, A.H. The stability of salt bridges at high temperatures: Implications for hyperthermophilic proteins 1 Edited by B. Honig. J. Mol. Biol. 1998, 284, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Liao, M.; Meng, X.; Somero, G.N. Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc. Natl. Acad. Sci. USA 2018, 115, 1274–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijma, H.J.; Floor, R.J.; Janssen, D.B. Structure- and sequence-analysis inspired engineering of proteins for enhanced thermostability. Curr. Opin. Struct. Biol. 2013, 23, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Cen, Y.K.; Zou, S.P.; Xue, Y.P.; Zheng, Y.G. Recent advances in the improvement of enzyme thermostability by structure modification. Crit. Rev. Biotechnol. 2020, 40, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.H.; Lin, M.C.; Lee, C.C.; Yu, S.M.; Wang, A.H.J.; Ho, T.-H.D. Enhancement of laccase activity by pre-incubation with organic solvents. Sci. Rep. 2019, 9, 9754. [Google Scholar] [CrossRef] [PubMed]
- Said, Z.S.A.A.M.; Arifi, F.A.M.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Leow, A.T.C.; Latip, W.; Ali, M.S.M. Unravelling protein -organic solvent interaction of organic solvent tolerant elastase from Pseudomonas aeruginosa strain K crystal structure. Int. J. Biol. Macromol. 2019, 127, 575–584. [Google Scholar] [CrossRef]
- Fitzpatrick, P.A.; Steinmetz, A.C.U.; Ringe, D.; Klibanov, A.M. Enzyme crystal structure in a neat organic solvent. Proc. Natl. Acad. Sci. USA 1993, 90, 8653–8657. [Google Scholar] [CrossRef] [Green Version]
- Cianci, M.; Tomaszewski, B.; Helliwell, J.R.; Halling, P.J. Crystallographic analysis of counterion effects on subtilisin enzymatic action in acetonitrile. J. Am. Chem. Soc. 2010, 132, 2293–2300. [Google Scholar] [CrossRef]
- English, A.C.; Groom, C.R.; Hubbard, R.E. Experimental and computational mapping of the binding surface of a crystalline protein. Protein Eng. Des. Sel. 2001, 14, 47–59. [Google Scholar] [CrossRef]
- Gupta, M.N.; Tyagi, R.; Sharma, S.; Karthikeyan, S.; Singh, T.P. Enhancement of catalytic efficiency of enzymes through exposure to anhydrous organic solvent at 70 °C. Three-dimensional structure of a treated serine proteinase at 2.2 Å resolution. Proteins Struct. Funct. Genet. 2000, 39, 226–234. [Google Scholar] [CrossRef]
- Schmitke, J.L.; Stern, L.J.; Klibanov, A.M. The crystal structure of subtilisin Carlsberg in anhydrous dioxane and its comparison with those in water and acetonitrile. Proc. Natl. Acad. Sci. USA 1997, 94, 4250–4255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepankova, V.; Damborsky, J.; Chaloupkova, R. Organic co-solvents affect activity, stability and enantioselectivity of haloalkane dehalogenases. Biotechnol. J. 2013, 8, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Polizzi, K.M.; Bommarius, A.S.; Broering, J.M.; Chaparro-Riggers, J.F. Stability of biocatalysts. Curr. Opin. Chem. Biol. 2007, 11, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Mohtashami, M.; Fooladi, J.; Haddad-Mashadrizeh, A.; Housaindokht, M.R.; Monhemi, H. Molecular mechanism of enzyme tolerance against organic solvents: Insights from molecular dynamics simulation. Int. J. Biol. Macromol. 2019, 122, 914–923. [Google Scholar] [CrossRef]
- Shao, Q. Methanol concentration dependent protein denaturing ability of guanidinium/methanol mixed solution. J. Phys. Chem. B 2014, 118, 6175–6185. [Google Scholar] [CrossRef]
- Pazhang, M.; Mardi, N.; Mehrnejad, F.; Chaparzadeh, N. The combinatorial effects of osmolytes and alcohols on the stability of pyrazinamidase: Methanol affects the enzyme stability through hydrophobic interactions and hydrogen bonds. Int. J. Biol. Macromol. 2018, 108, 1339–1347. [Google Scholar] [CrossRef]
- Kuper, J.; Wong, T.S.; Roccatano, D.; Wilmanns, M.; Schwaneberg, U. Understanding a mechanism of organic cosolvent inactivation in heme monooxygenase P450 BM-3. J. Am. Chem. Soc. 2007, 129, 5786–5787. [Google Scholar] [CrossRef]
- Roccatano, D. Computer simulations study of biomolecules in non-aqueous or cosolvent/water mixture solutions. Curr. Protein Pept. Sci. 2008, 9, 407–426. [Google Scholar] [CrossRef]
- Srivastava, K.R.; Goyal, B.; Kumar, A.; Durani, S. Scrutiny of electrostatic-driven conformational ordering of polypeptide chains in DMSO: A study with a model oligopeptide. RSC Adv. 2017, 7, 27981–27991. [Google Scholar] [CrossRef]
- Johnson, M.E.; Malardier-Jugroot, C.; Head-Gordon, T. Effects of co-solvents on peptide hydration water structure and dynamics. Phys. Chem. Chem. Phys. 2010, 12, 393–405. [Google Scholar] [CrossRef] [Green Version]
- Markel, U.; Zhu, L.; Frauenkron-Machedjou, V.; Zhao, J.; Bocola, M.; Davari, M.; Jaeger, K.-E.; Schwaneberg, U. Are directed evolution approaches efficient in exploring nature’s potential to stabilize a lipase in organic cosolvents? Catalysts 2017, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Nie, K.; Xu, H.; Xiong, X.; Krastev, R.; Wang, F.; Tan, T.; Liu, L. Insight into the mechanism behind the activation phenomenon of lipase from Thermus thermophilus HB8 in polar organic solvents. J. Mol. Catal. B Enzym. 2016, 133, S400–S409. [Google Scholar] [CrossRef]
- Kamal, M.Z.; Yedavalli, P.; Deshmukh, M.V.; Rao, N.M. Lipase in aqueous-polar organic solvents: Activity, structure, and stability. Protein Sci. 2013, 22, 904–915. [Google Scholar] [CrossRef] [Green Version]
- Batra, R.; Gupta, M.N. Enhancement of enzyme activity in aqueous-organic solvent mixtures. Biotechnol. Lett. 1994, 16, 1059–1064. [Google Scholar] [CrossRef]
- Kudryashova, E.V.; Gladilin, A.K.; Vakurov, A.V.; Heitz, F.; Levashov, A.V.; Mozhaev, V.V. Enzyme-polyelectrolyte complexes in water-ethanol mixtures: Negatively charged groups artificially introduced into α-chymotrypsin provide additional activation and stabilization effects. Biotechnol. Bioeng. 1997, 55, 267–277. [Google Scholar] [CrossRef]
- Gord Noshahri, N.; Fooladi, J.; Syldatk, C.; Engel, U.; Heravi, M.M.; Zare Mehrjerdi, M.; Rudat, J. Screening and comparative characterization of microorganisms from iranian soil samples showing ω-transaminase activity toward a plethora of substrates. Catalysts 2019, 9, 874. [Google Scholar] [CrossRef] [Green Version]
- Sirotkin, V.A.; Zinatullin, A.N.; Solomonov, B.N.; Faizullin, D.A.; Fedotov, V.D. Calorimetric and Fourier transform infrared spectroscopic study of solid proteins immersed in low water organic solvents. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2001, 1547, 359–369. [Google Scholar] [CrossRef]
- Sirotkin, V.A.; Kuchierskaya, A.A. α-chymotrypsin in water-acetone and water-dimethyl sulfoxide mixtures: Effect of preferential solvation and hydration. Proteins. Struct. Funct. Bioinforma. 2017, 85, 1808–1819. [Google Scholar] [CrossRef]
- Bezsudnova, E.Y.; Boyko, K.M.; Nikolaeva, A.Y.; Zeifman, Y.S.; Rakitina, T.V.; Suplatov, D.A.; Popov, V.O. Biochemical and structural insights into PLP fold type IV transaminase from Thermobaculum terrenum. Biochimie 2019, 158, 130–138. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Lakowicz, J.R., Ed.; Springer US: Boston, MA, USA, 2006; ISBN 978-0-387-31278-1. [Google Scholar]
- Bushueva, T.L.; Busel, E.P.; Burstein, E.A. Relationship of thermal quenching of protein fluorescence to intramolecular structural mobility. Biochim. Biophys. Acta Protein Struct. 1978, 534, 141–152. [Google Scholar] [CrossRef]
- Boyko, K.M.; Stekhanova, T.N.; Nikolaeva, A.Y.; Mardanov, A.V.; Rakitin, A.L.; Ravin, N.V.; Bezsudnova, E.Y.; Popov, V.O. First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for l-amino acids and R-amines. Extremophiles 2016, 20, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, X.; Engel, P.C. The specificity and kinetic mechanism of branched-chain amino acid aminotransferase from Escherichia coli studied with a new improved coupled assay procedure and the enzyme’s potential for biocatalysis. FEBS J. 2014, 281, 391–400. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Burstein, E.A. Some aspects of studies of thermal transitions in proteins by means of their intrinsic fluorescence. Biophys. Chem. 1984, 19, 265–271. [Google Scholar] [CrossRef]
- Available online: http://cib.cf.ocha.ac.jp/bitool/HBOND (accessed on 29 April 2009).
- Bezsudnova, E.Y.; Popov, V.O.; Boyko, K.M. Structural insight into the substrate specificity of PLP fold type IV transaminases. Appl. Microbiol. Biotechnol. 2020, 104, 2343–2357. [Google Scholar] [CrossRef] [PubMed]
- Botero, L.M.; Brown, K.B.; Brumefield, S.; Burr, M.; Castenholz, R.W.; Young, M.; McDermott, T.R. Thermobaculum terrenum gen. nov., sp. nov.: A non-phototrophic gram-positive thermophile representing an environmental clone group related to the Chloroflexi (green non-sulfur bacteria) and Thermomicrobia. Arch. Microbiol. 2004, 181, 269–277. [Google Scholar] [CrossRef]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef] [PubMed]






| Solvent | Vmax, U/mg | Km, mM | kcat/Km, s−1 M−1 |
|---|---|---|---|
| Buffer | 178 ± 23 | 7.8 ± 2.3 | 13,700 ± 4400 |
| 15% DMSO | 280 ± 40 | 12 ± 4 | 14,000 ± 5000 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bezsudnova, E.Y.; Nikolaeva, A.Y.; Kleymenov, S.Y.; Petrova, T.E.; Zavialova, S.A.; Tugaeva, K.V.; Sluchanko, N.N.; Popov, V.O. Counterbalance of Stability and Activity Observed for Thermostable Transaminase from Thermobaculum terrenum in the Presence of Organic Solvents. Catalysts 2020, 10, 1024. https://doi.org/10.3390/catal10091024
Bezsudnova EY, Nikolaeva AY, Kleymenov SY, Petrova TE, Zavialova SA, Tugaeva KV, Sluchanko NN, Popov VO. Counterbalance of Stability and Activity Observed for Thermostable Transaminase from Thermobaculum terrenum in the Presence of Organic Solvents. Catalysts. 2020; 10(9):1024. https://doi.org/10.3390/catal10091024
Chicago/Turabian StyleBezsudnova, Ekaterina Yu., Alena Yu. Nikolaeva, Sergey Y. Kleymenov, Tatiana E. Petrova, Sofia A. Zavialova, Kristina V. Tugaeva, Nikolai N. Sluchanko, and Vladimir O. Popov. 2020. "Counterbalance of Stability and Activity Observed for Thermostable Transaminase from Thermobaculum terrenum in the Presence of Organic Solvents" Catalysts 10, no. 9: 1024. https://doi.org/10.3390/catal10091024
APA StyleBezsudnova, E. Y., Nikolaeva, A. Y., Kleymenov, S. Y., Petrova, T. E., Zavialova, S. A., Tugaeva, K. V., Sluchanko, N. N., & Popov, V. O. (2020). Counterbalance of Stability and Activity Observed for Thermostable Transaminase from Thermobaculum terrenum in the Presence of Organic Solvents. Catalysts, 10(9), 1024. https://doi.org/10.3390/catal10091024