Functional Characterization of Recombinant Raw Starch Degrading α-Amylase from Roseateles terrae HL11 and Its Application on Cassava Pulp Saccharification
Abstract
:1. Introduction
2. Results
2.1. Amylase Gene Identification from R. terrae and Sequence Analysis
2.2. Heterologous Expression and Purification of Amylase
2.3. Biochemical Characterization
2.4. Cleavage Pattern on Maltooligosaccharides
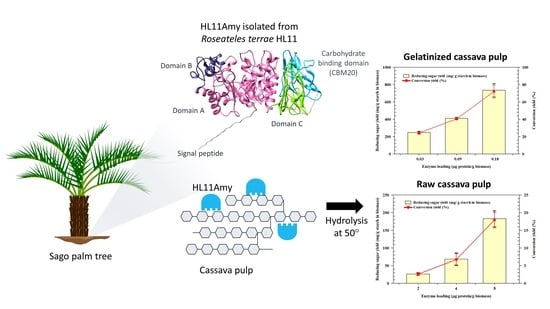
2.5. Hydrolysis of Cassava Pulp
3. Discussion
4. Materials and Methods
4.1. Chemicals, Bacterial Strains and Plasmids
4.2. Identification of HL11Amy Amylase Gene from R. terrae HL11
4.3. Heterologous Expression and Purification of Amylase
4.4. Purification of Recombinant HL11Amy
4.5. Enzyme Activity Assay
4.6. Determination of Biochemical Properties of Recombinant HL11Amy
4.7. Determination of Kinetic Parameters (Vmax and Km) on Various Substrates
4.8. Analysis of Cleavage Pattern on Maltooligosaccharides
4.9. Hydrolysis of Raw and Gelatinized Cassava Pulp
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Food and agriculture organization of the united nations. Food Outlook: Biannual report on global food markets 2016. In Food Outlook; Food and Agriculture Organization of the United Nations: Rome, Italy, 2016. [Google Scholar]
- Sowcharoensuk, C. Thailand Industry Outlook 2020–2022; Krungsri Research: Bangkok, Thailand, 2020. [Google Scholar]
- Ghimire, A.; Sen, R.; Annachhatre, A.P. Biosolid management options in cassava starch industries of Thailand: Present practice and future possibilities. Procedia Chem. 2015, 14, 66–75. [Google Scholar] [CrossRef] [Green Version]
- Trakulvichean, S.; Chaiprasert, P.; Otmakhova, J.; Songkasiri, W. Comparison of fermented animal feed and mushroom growth media as two value-added options for waste cassava pulp management. Waste Manag. Res. 2017, 35, 1210–1219. [Google Scholar] [CrossRef]
- Bunterngsook, B.; Laothanachareon, T.; Natrchalayuth, S.; Lertphanich, S.; Fujii, T.; Inoue, H.; Youngthong, C.; Chantasingh, D.; Eurwilaichitr, L.; Champreda, V. Optimization of a minimal synergistic enzyme system for hydrolysis of raw cassava pulp. RSC Adv. 2017, 7, 48444–48453. [Google Scholar] [CrossRef] [Green Version]
- Hermiati, E.; Azuma, J.-i.; Mangunwidjaja, D.; Sunarti, T.C.; Suparno, O.; Prasetya, B. Hydrolysis of carbohydrates in cassava pulp and tapioca flour under microwave irradiation. Indones. J. Chem. 2011, 11, 238–245. [Google Scholar] [CrossRef]
- Leite, A.; Zanon, C.D.; Menegalli, F.C. Isolation and characterization of cellulose nanofibers from cassava root bagasse and peelings. Carbohydr. Polym. 2017, 157, 962–970. [Google Scholar] [CrossRef]
- Widiarto, S.; Pramono, E.; Rochliadi, A.; Arcana, I.M. Cellulose nanofibers preparation from cassava peels via mechanical disruption. Fibers 2019, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Janecek, S.; Gabrisko, M. Remarkable evolutionary relatedness among the enzymes and proteins from the alpha-amylase family. Cell. Mol. Life Sci. 2016, 73, 2707–2725. [Google Scholar] [CrossRef]
- Janecek, S.; Kuchtova, A. In silico identification of catalytic residues and domain fold of the family GH119 sharing the catalytic machinery with the alpha-amylase family GH57. FEBS Lett. 2012, 586, 3360–3366. [Google Scholar] [CrossRef] [Green Version]
- Janeček, Š.; Svensson, B. How many α-amylase GH families are there in the CAZy database? Amylase 2022, 6, 1–10. [Google Scholar] [CrossRef]
- Janecek, S.; Svensson, B.; MacGregor, E.A. alpha-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef]
- Janecek, S.; Zamocka, B. A new GH13 subfamily represented by the alpha-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles 2020, 24, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Janíčková, Z.; Janeček, Š. In silico analysis of fungal and chloride-dependent α-amylases within the family GH13 with identification of possible secondary surface-binding sites. Molecules 2021, 26, 5704. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarian, F.D.; Janecek, S.; Pijning, T.; Nurachman, Z.; Radjasa, O.K.; Dijkhuizen, L.; Natalia, D.; van der Maarel, M.J. A new group of glycoside hydrolase family 13 alpha-amylases with an aberrant catalytic triad. Sci. Rep. 2017, 7, 44230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stam, M.R.; Danchin, E.G.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of alpha-amylase-related proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef] [Green Version]
- Božić, N.; Lončar, N.; Slavić, M.Š.; Vujčić, Z. Raw starch degrading α-amylases: An unsolved riddle. Amylase 2017, 1, 12–25. [Google Scholar] [CrossRef]
- Hedin, N.; Velazquez, M.B.; Barchiesi, J.; Gomez-Casati, D.F.; Busi, M.V. CBM20CP, a novel functional protein of starch metabolism in green algae. Plant Mol. Biol. 2022, 108, 363–378. [Google Scholar] [CrossRef]
- Janecek, S.; Marecek, F.; MacGregor, E.A.; Svensson, B. Starch-binding domains as CBM families-history, occurrence, structure, function and evolution. Biotechnol. Adv. 2019, 37, 107451. [Google Scholar] [CrossRef]
- Gohel, V.; Duan, G. Conventional process for ethanol production from Indian broken rice and pearl millet. Bioprocess Biosyst. Eng. 2012, 35, 1297–1308. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, P.; Ge, X.; Xia, Y.; Hao, Z.; Liu, J.; Peng, M. Recent advances in microbial raw starch degrading enzymes. Appl. Biochem. Biotechnol. 2010, 160, 988–1003. [Google Scholar] [CrossRef]
- Serin, B.; Akcan, N.; Uyar, F. Production and optimization of α-amylase from Bacillus circulans ATCC 4516 with solid state fermentation. J. Biol. Chem. 2012, 40, 393–400. [Google Scholar]
- Gomila, M.; Bowien, B.; Falsen, E.; Moore, E.R.; Lalucat, J. Description of Roseateles aquatilis sp. nov. and Roseateles terrae sp. nov., in the class Betaproteobacteria, and emended description of the genus Roseateles. Int. J. Syst. Evol. Microbiol. 2008, 58, 6–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, A.A.; Eguchi, T.; Mayumi, D.; Kato, S.; Shintani, N.; Kamini, N.R.; Nakajima-Kambe, T. Purification and properties of novel aliphatic-aromatic co-polyesters degrading enzymes from newly isolated Roseateles depolymerans strain TB-87. Polym. Degrad. Stab. 2013, 98, 609–618. [Google Scholar] [CrossRef]
- Suyama, T.; Shigematsu, T.; Takaichi, S.; Nodasaka, Y.; Fujikawa, S.; Hosoya, H.; Tokiwa, Y.; Kanagawa, T.; Hanada, S. Roseateles depolymerans gen. nov., sp. nov., a new bacteriochlorophyll a-containing obligate aerobe belonging to the beta-subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 1999, 49, 449–457. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Svensson, B.; Jespersen, H.; Sierks, M.R.; MacGregor, E.A. Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem. J. 1989, 264, 309–311. [Google Scholar] [CrossRef] [Green Version]
- Christiansen, C.; Abou Hachem, M.; Janecek, S.; Vikso-Nielsen, A.; Blennow, A.; Svensson, B. The carbohydrate-binding module family 20--diversity, structure, and function. FEBS J. 2009, 276, 5006–5029. [Google Scholar] [CrossRef]
- Janecek, S. Sequence similarities and evolutionary relationships of microbial, plant and animal alpha-amylases. Eur. J. Biochem. 1994, 224, 519–524. [Google Scholar] [CrossRef]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structural basis of alpha-amylase activation by chloride. Protein Sci. 2002, 11, 1435–1441. [Google Scholar] [CrossRef]
- Feller, G.; Bussy, O.; Houssier, C.; Gerday, C. Structural and functional aspects of chloride binding to Alteromonas haloplanctis alpha-amylase. J. Biol. Chem. 1996, 271, 23836–23841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feller, G.; Lonhienne, T.; Deroanne, C.; Libioulle, C.; Van Beeumen, J.; Gerday, C. Purification, characterization, and nucleotide sequence of the thermolabile alpha-amylase from the antarctic psychrotroph Alteromonas haloplanctis A23. J. Biol. Chem. 1992, 267, 5217–5221. [Google Scholar] [CrossRef]
- Coronado, M.A.; Vargas, C.; Mellado, E.; Tegos, G.; Drainas, C.; Nieto, J.N.J.; Ventosa, A. The alpha-amylase gene amyH of the moderate halophile Halomonas meridiana: Cloning and molecular characterization. Microbiology 2000, 146, 861–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.-S.; Na, H.-K.; Jhon, D.-Y.; Yoo, O.J.; Chun, S.-B.; Wui, I.-S. Cloning, sequencing and expression of the amylase isozyme gene from Pseudomonas sp. KFCC 10818. Biotechnol. Lett. 1996, 18, 169–174. [Google Scholar] [CrossRef]
- Petříček, M.; Tichý, P.; Kuncová, M. Characterization of the α-amylase-encoding gene from Thermomonospora curvata. Gene 1992, 112, 77–83. [Google Scholar] [CrossRef]
- Yang, C.H.; Liu, W.H. Cloning and characterization of a maltotriose-producing alpha-amylase gene from Thermobifida fusca. J. Ind. Microbiol. Biotechnol. 2007, 34, 325–330. [Google Scholar] [CrossRef]
- Machius, M.; Declerck, N.; Huber, R.; Wiegand, G. Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad. Structure 1998, 6, 281–292. [Google Scholar] [CrossRef] [Green Version]
- Da Lage, J.-L.; Feller, G.; Janecek, S. Horizontal gene transfer from Eukarya to bacteria and domain shuffling: The α-amylase model. Cell. Mol. Life Sci. 2004, 61, 97–109. [Google Scholar] [CrossRef]
- Da Lage, J.L.; Binder, M.; Hua-Van, A.; Janeček, S.; Casane, D. Gene make-up: Rapid and massive intron gains after horizontal transfer of a bacterial α-amylase gene to Basidiomycetes. BMC Evol. Biol. 2013, 13, 40. [Google Scholar] [CrossRef] [Green Version]
- Mehta, D.; Satyanarayana, T. Domain C of thermostable alpha-amylase of Geobacillus thermoleovorans mediates raw starch adsorption. Appl. Microbiol. Biotechnol. 2014, 98, 4503–4519. [Google Scholar] [CrossRef]
- Sahnoun, M.; Jemli, S.; Trabelsi, S.; Ayadi, L.; Bejar, S. Aspergillus Oryzae S2 alpha-amylase domain C involvement in activity and specificity: In vivo proteolysis, molecular and docking studies. PLoS ONE 2016, 11, e0153868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockburn, D.; Nielsen, M.M.; Christiansen, C.; Andersen, J.M.; Rannes, J.B.; Blennow, A.; Svensson, B. Surface binding sites in amylase have distinct roles in recognition of starch structure motifs and degradation. Int. J. Biol. Macromol. 2015, 75, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Baroroh, U.; Yusuf, M.; Rachman, S.D.; Ishmayana, S.; Syamsunarno, M.; Levita, J.; Subroto, T. The importance of surface-binding site towards starch-adsorptivity level in alpha-amylase: A review on structural point of view. Enzyme Res. 2017, 2017, 4086845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tawil, G.; Vikso-Nielsen, A.; Rolland-Sabate, A.; Colonna, P.; Buleon, A. Hydrolysis of concentrated raw starch: A new very efficient alpha-amylase from Anoxybacillus flavothermus. Carbohydr. Polym. 2012, 87, 46–52. [Google Scholar] [CrossRef]
- Peng, H.; Zheng, Y.; Chen, M.; Wang, Y.; Xiao, Y.; Gao, Y. A starch-binding domain identified in alpha-amylase (AmyP) represents a new family of carbohydrate-binding modules that contribute to enzymatic hydrolysis of soluble starch. FEBS Lett. 2014, 588, 1161–1167. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez Sanoja, R.; Morlon-Guyot, J.; Jore, J.; Pintado, J.; Juge, N.; Guyot, J.P. Comparative characterization of complete and truncated forms of Lactobacillus amylovorus alpha-amylase and role of the C-terminal direct repeats in raw-starch binding. Appl. Environ. Microbiol. 2000, 66, 3350–3356. [Google Scholar] [CrossRef] [Green Version]
- Sumitani, J.; Tottori, T.; Kawaguchi, T.; Arai, M. New type of starch-binding domain: The direct repeat motif in the C-terminal region of Bacillus sp. no. 195 alpha-amylase contributes to starch binding and raw starch degrading. Biochem. J. 2000, 350, 477–484. [Google Scholar] [CrossRef]
- Peng, H.; Li, R.; Li, F.; Zhai, L.; Zhang, X.; Xiao, Y.; Gao, Y. Extensive hydrolysis of raw rice starch by a chimeric alpha-amylase engineered with alpha-amylase (AmyP) and a starch-binding domain from Cryptococcus sp. S-2. Appl. Microbiol. Biotechnol. 2018, 102, 743–750. [Google Scholar] [CrossRef]
- Liu, X.; Li, L.; Yu, J.; Copeland, L.; Wang, S.; Wang, S. In vitro digestibility of starches with different crystalline polymorphs at low alpha-amylase activity to substrate ratio. Food Chem. 2021, 349, 129170. [Google Scholar] [CrossRef]
- Vidilaseris, K.; Hidayat, K.; Retnoningrum, D.S.; Nurachman, Z.; Noer, A.S.; Natalia, D. Biochemical characterization of a raw starch degrading α-amylase from the Indonesian marine bacterium Bacillus sp. ALSHL3. Biologia 2009, 64, 1047–1052. [Google Scholar] [CrossRef]
- Gangadharan, D.; Nampoothiri, K.M.; Sivaramakrishnan, S.; Pandey, A. Biochemical characterization of raw-starch-digesting alpha amylase purified from Bacillus amyloliquefaciens. Appl. Biochem. Biotechnol. 2009, 158, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Peng, H.; Wang, Y.; Liu, Y.; Han, F.; Xiao, Y.; Gao, Y. Preferential and rapid degradation of raw rice starch by an alpha-amylase of glycoside hydrolase subfamily GH13_37. Appl. Microbiol. Biotechnol. 2012, 94, 1577–1584. [Google Scholar] [CrossRef] [PubMed]
- Maktouf, S.; Kamoun, A.; Moulis, C.; Remaud-Simeon, M.; Ghribi, D.; Chaabouni, S.E. A new raw-starch-digesting alpha-amylase: Production under solid-state fermentation on crude millet and biochemical characterization. J. Microbiol. Biotechnol. 2013, 23, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivlata, L.; Satyanarayana, T. Characteristics of raw starch-digesting alpha-amylase of Streptomyces badius DB-1 with transglycosylation activity and its applications. Appl. Biochem. Biotechnol. 2017, 181, 1283–1303. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Xue, S.; Deng, P.; Zhang, X.; Wang, X.; Xiao, Y.; Fang, Z. AmyZ1: A novel alpha-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches. Biotechnol. Biofuels 2019, 12, 95. [Google Scholar] [CrossRef]
- Li, Z.; Wu, J.; Zhang, B.; Wang, F.; Ye, X.; Huang, Y.; Huang, Q.; Cui, Z. AmyM, a novel maltohexaose-forming alpha-amylase from Corallococcus sp. strain EGB. Appl. Environ. Microbiol. 2015, 81, 1977–1987. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.W.; Kramhoft, B.; Bozonnet, S.; Abou Hachem, M.; Stipp, S.L.; Svensson, B.; Willemoes, M. Degradation of the starch components amylopectin and amylose by barley alpha-amylase 1: Role of surface binding site 2. Arch. Biochem. Biophys. 2012, 528, 1–6. [Google Scholar] [CrossRef]
- Nielsen, M.M.; Bozonnet, S.; Seo, E.S.; Motyan, J.A.; Andersen, J.M.; Dilokpimol, A.; Abou Hachem, M.; Gyemant, G.; Naested, H.; Kandra, L.; et al. Two secondary carbohydrate binding sites on the surface of barley alpha-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry 2009, 48, 7686–7697. [Google Scholar] [CrossRef]
- Edwards, C.H.; Maillot, M.; Parker, R.; Warren, F.J. A comparison of the kinetics of in vitro starch digestion in smooth and wrinkled peas by porcine pancreatic alpha-amylase. Food Chem. 2018, 244, 386–393. [Google Scholar] [CrossRef] [Green Version]
- Martens, B.M.J.; Gerrits, W.J.J.; Bruininx, E.; Schols, H.A. Amylopectin structure and crystallinity explains variation in digestion kinetics of starches across botanic sources in an in vitro pig model. J. Anim. Sci. Biotechnol. 2018, 9, 91. [Google Scholar] [CrossRef] [Green Version]
- Sarian, F.D.; van der Kaaij, R.M.; Kralj, S.; Wijbenga, D.J.; Binnema, D.J.; van der Maarel, M.J.; Dijkhuizen, L. Enzymatic degradation of granular potato starch by Microbacterium aurum strain B8.A. Appl. Microbiol. Biotechnol. 2012, 93, 645–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yu, J.; Li, F.; Peng, H.; Zhang, X.; Xiao, Y.; He, C. Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13. Sci. Rep. 2017, 7, 44067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogunsona, E.; Ojogbo, E.; Mekonnen, T. Advanced material applications of starch and its derivatives. Eur. Polym. J. 2018, 108, 570–581. [Google Scholar] [CrossRef]
- Gallant, D.J.; Bouchet, B.; Buleon, A.; Perez, S. Physical characteristics of starch granules and susceptibility to enzymatic degradation. Eur. J. Clin. Nutr. 1992, 46, S3–S16. [Google Scholar]
- Shofiyah, S.S.; Yuliani, D.; Widya, N.; Sarian, F.D.; Puspasari, F.; Radjasa, O.K.; Natalia, D. Isolation, expression, and characterization of raw starch degrading alpha-amylase from a marine lake Bacillus megaterium NL3. Heliyon 2020, 6, e05796. [Google Scholar] [CrossRef]
- Dome, K.; Podgorbunskikh, E.; Bychkov, A.; Lomovsky, O. Changes in the crystallinity degree of starch having different types of crystal structure after mechanical pretreatment. Polymers 2020, 12, 641. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Zhang, L.; Cai, X.; Liu, Q.; Zhang, C.; Wei, C. The relationship between enzyme hydrolysis and the components of rice starches with the same genetic background and amylopectin structure but different amylose contents. Food Hydrocoll. 2018, 84, 406–413. [Google Scholar] [CrossRef]
- Alcázar-Alay, S.C.; Meireles, M.A.A. Physicochemical properties, modifications and applications of starches from different botanical sources. Food Sci. Technol. 2015, 35, 215–236. [Google Scholar] [CrossRef] [Green Version]
- Hii, S.L.; Tan, J.S.; Ling, T.C.; Ariff, A.B. Pullulanase: Role in starch hydrolysis and potential industrial applications. Enzyme Res. 2012, 2012, 921362. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Zhang, L.; Dai, Y.; Yu, J. Physicochemical properties of starch obtained from Dioscorea nipponica Makino comparison with other tuber starches. J. Food Eng. 2007, 82, 436–442. [Google Scholar] [CrossRef]
- Bai, Y.; Huang, H.; Meng, K.; Shi, P.; Yang, P.; Luo, H.; Luo, C.; Feng, Y.; Zhang, W.; Yao, B. Identification of an acidic α-amylase from Alicyclobacillus sp. A4 and assessment of its application in the starch industry. Food Chem. 2012, 131, 1473–1478. [Google Scholar] [CrossRef]
- Djuma’ali, D.a.; Soewarno, N.; Sumarno, S.; Primarini, D.; Sumaryono, W. Cassava pulp as a biofuel feedstock of an enzymatic hydrolysis process. Makara J. Technol. 2011, 15, 183–192. [Google Scholar] [CrossRef] [Green Version]
- Virunanon, C.; Ouephanit, C.; Burapatana, V.; Chulalaksananukul, W. Cassava pulp enzymatic hydrolysis process as a preliminary step in bio-alcohols production from waste starchy resources. J. Clean. Prod. 2013, 39, 273–279. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 2002, 31, 426–428. [Google Scholar] [CrossRef]
- Peng, H.; Chen, M.; Yi, L.; Zhang, X.; Wang, M.; Xiao, Y.; Zhang, N. Identification and characterization of a novel raw-starch-degrading α-amylase (AmyASS) from the marine fish pathogen Aeromonas salmonicida ssp. salmonicida. J. Mol. Catal. B Enzym. 2015, 119, 71–77. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Smith, T.J. SusG: A Unique cell-membrane-associated α-amylase from a prominent human gut symbiont targets complex starch molecules. Structure 2010, 18, 200–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puspasari, F.; Radjasa, O.K.; Noer, A.S.; Nurachman, Z.; Syah, Y.M.; van der Maarel, M.; Dijkhuizen, L.; Janeček, Š.; Natalia, D. Raw starch–degrading α-amylase from Bacillus aquimaris MKSC 6.2: Isolation and expression of the gene, bioinformatics and biochemical characterization of the recombinant enzyme. J. Appl. Microbiol. 2013, 114, 108–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, J.K.; Borah, A.; Mahanta, C.L.; Mukherjee, A.K. Cloning and overexpression of raw starch digesting α-amylase gene from Bacillus subtilis strain AS01a in Escherichia coli and application of the purified recombinant α-amylase (AmyBS-I) in raw starch digestion and baking industry. J. Mol. Catal. B Enzym. 2013, 97, 118–129. [Google Scholar] [CrossRef]
- Attanayaka, D.P.S.T.G.; Silva, S.N.T.D.; Nirosha, S.F.; Aththanayaka, A.M.W.S. Isolation of raw starch hydrolysing fungi and purification of α-amylase from Geotrichum candidum CMSS06. J. Natl. Sci. Found. Sri. Lanka 2009, 37, 93–98. [Google Scholar] [CrossRef]
- Tawil, G.; Viksø-Nielsen, A.; Rolland-Sabaté, A.; Colonna, P.; Buléon, A. In depth study of a new highly efficient raw starch hydrolyzing α-amylase from Rhizomucor sp. Biomacromolecules 2011, 12, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Hostinová, E.; Janeček, Š.; Gašperík, J. Gene Sequence, Bioinformatics and enzymatic characterization of α-amylase from Saccharomycopsis fibuligera KZ. Protein J. 2010, 29, 355–364. [Google Scholar] [CrossRef]







| Substrates | Gelatinized Substrates | Raw Substrates | ||
|---|---|---|---|---|
| Relative Activity (%) | Specific Activity (U/mg) | Relative Activity (%) | Specific Activity (U/mg) | |
| Soluble starch | 100 ± 1.8 | 6270 ± 70 | 100 ± 3.5 | 1030 ± 40 |
| Amylose from potato | 147 ± 2.6 | 8890 ± 160 | 531 ± 1.7 | 5470 ± 20 |
| Amypectin from potato | 174 ± 4.1 | 10,530 ± 250 | 463 ± 5.7 | 4770 ± 60 |
| Cassava starch | 166 ± 5.9 | 10,000 ± 360 | 8.44 ± 0.75 | 90 ± 8 |
| Potato starch | 175 ± 4.8 | 10,570 ± 290 | 13.0 ± 0.75 | 130 ± 8 |
| Rice starch | 169 ± 4.2 | 10,220 ± 250 | 191 ± 0.83 | 1970 ± 9 |
| Cassava pulp | 103 ± 3.2 | 6200 ± 190 | 0.93 ± 0.04 | 10 ± 0.4 |
| Pullulan | 3.58 ± 0.7 | 220 ± 40 | ND a | ND |
| Soluble Starch | Amylose | Amylopectin | Cassava Starch | Potato Starch | Rice Starch | |
|---|---|---|---|---|---|---|
| Gelatinized substrates | ||||||
| Km (mg/mL) | 4.82 ± 0.86 | 2.40 ± 0.54 | 5.01 ± 0.43 | 5.17 ± 1.40 | 3.84 ± 0.65 | 5.38 ± 0.36 |
| Vmax (U/mg) | 19,800 ± 1650 | 5660 ± 460 | 10,750 ± 430 | 11,670 ± 1510 | 8660 ± 630 | 16,620 ± 550 |
| kcat (1/s) | 25,720 | 7350 | 13,960 | 15,170 | 11,250 | 21,590 |
| kcat/Km (mL/mg·sec) | 5330 | 3050 | 2780 | 2930 | 2930 | 4010 |
| Raw substrates | ||||||
| Km (mg/mL) | 13.51 ± 3.73 | 12.42 ± 2.08 | 4.28 ± 1.41 | 40.88 ± 2.58 | 30.88 ± 7.44 | 19.0 ± 3.00 |
| Vmax (U/mg) | 800 ± 15.6 | 6050 ± 650 | 6120 ± 900 | 2.66 ± 0.30 | 560 ± 0.66 | 4880 ± 1100 |
| kcat (1/s) | 1040 | 7860 | 7950 | 3.46 | 720 | 6340 |
| kcat/Km (mL/mg·sec) | 76.9 | 630 | 1850 | 0.08 | 23.4 | 330 |
| Starch | Granule Shape | Granule Size (µM) | Amylose Content (%) | Amylopectin Content (%) | Pasting Temperature (°C) | Crystalline Type | Degree of Crystallinity (%) |
|---|---|---|---|---|---|---|---|
| Cassava starch | Round with a truncated end | 5–35 | 19.49 | 83 | 62–72 | C | 48.0 |
| Potato starch | Spherical or oval | 5–100 | 21 | 79 | 52–72 | B | 45.9 |
| Rice starch | Polygonal, angular | 3–8 | 17 | 83 | 55–79 | A | 30.7–35.7 |
| Amylose from potato | - | - | 98 | - | - | - | - |
| Amylopectin from potato | - | - | - | 98 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prongjit, D.; Lekakarn, H.; Bunterngsook, B.; Aiewviriyasakul, K.; Sritusnee, W.; Champreda, V. Functional Characterization of Recombinant Raw Starch Degrading α-Amylase from Roseateles terrae HL11 and Its Application on Cassava Pulp Saccharification. Catalysts 2022, 12, 647. https://doi.org/10.3390/catal12060647
Prongjit D, Lekakarn H, Bunterngsook B, Aiewviriyasakul K, Sritusnee W, Champreda V. Functional Characterization of Recombinant Raw Starch Degrading α-Amylase from Roseateles terrae HL11 and Its Application on Cassava Pulp Saccharification. Catalysts. 2022; 12(6):647. https://doi.org/10.3390/catal12060647
Chicago/Turabian StyleProngjit, Daran, Hataikarn Lekakarn, Benjarat Bunterngsook, Katesuda Aiewviriyasakul, Wipawee Sritusnee, and Verawat Champreda. 2022. "Functional Characterization of Recombinant Raw Starch Degrading α-Amylase from Roseateles terrae HL11 and Its Application on Cassava Pulp Saccharification" Catalysts 12, no. 6: 647. https://doi.org/10.3390/catal12060647
APA StyleProngjit, D., Lekakarn, H., Bunterngsook, B., Aiewviriyasakul, K., Sritusnee, W., & Champreda, V. (2022). Functional Characterization of Recombinant Raw Starch Degrading α-Amylase from Roseateles terrae HL11 and Its Application on Cassava Pulp Saccharification. Catalysts, 12(6), 647. https://doi.org/10.3390/catal12060647