Application of Cell-Free Protein Synthesis for Faster Biocatalyst Development
Abstract
:1. Introduction
2. Cell-Free Protein Synthesis
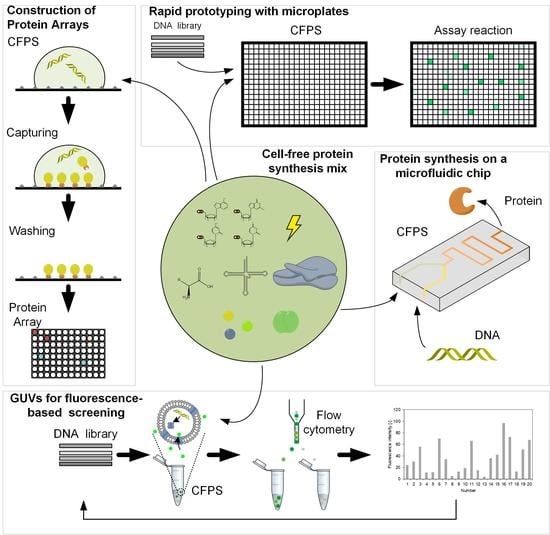
3. Applications of Cell-Free Protein Synthesis
4. Limitations and Challenges of CFPS
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Schmid, A.; Dordick, J.S.; Hauer, B.; Kiener, A.; Wubbolts, M.; Witholt, B. Industrial biocatalysis today and tomorrow. Nature 2001, 409, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Pellis, A.; Cantone, S.; Ebert, C.; Gardossi, L. Evolving biocatalysis to meet bioeconomy challenges and opportunities. New Biotechnol. 2018, 40, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, K.; Lütz, S. Recent developments and challenges of biocatalytic processes in the pharmaceutical industry. Curr. Opin. Green Sustain. Chem. 2018, 11, 58–64. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. The fourth wave of biocatalysis is approaching. Philos. Trans. R. Soc. A 2018, 376. [Google Scholar] [CrossRef]
- Schmitz, L.M.; Rosenthal, K.; Lütz, S. Enzyme-based electrobiotechnological synthesis. In Bioelectrosynthesis. Advances in Biochemical Engineering/Biotechnology; Harnisch, F., Holtmann, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 87–134. [Google Scholar]
- Wang, X.D.; Saba, T.; Yiu, H.H.P.; Howe, R.F.; Anderson, J.A.; Shi, J.F. Cofactor NAD(P)H regeneration inspired by heterogeneous pathways. Chem-Us 2017, 2, 621–654. [Google Scholar] [CrossRef]
- Karande, R.; Schmid, A.; Buehler, K. Applications of multiphasic microreactors for biocatalytic reactions. Org. Process. Res. Dev. 2016, 20, 361–370. [Google Scholar] [CrossRef]
- Weissman, S.A.; Anderson, N.G. Design of experiments (DoE) and process optimization. A review of recent publications. Org. Process. Res. Dev. 2015, 19, 1605–1633. [Google Scholar] [CrossRef]
- Gernaey, K.V.; Lantz, A.E.; Tufvesson, P.; Woodley, J.M.; Sin, G. Application of mechanistic models to fermentation and biocatalysis for next-generation processes. Trends Biotechnol. 2010, 28, 346–354. [Google Scholar] [CrossRef]
- Fernandes, P. Miniaturization in biocatalysis. Int. J. Mol. Sci. 2010, 11, 858–879. [Google Scholar] [CrossRef] [PubMed]
- Wohlgemuth, R. Biocatalytic process design and reaction engineering. Chem. Biochem. Eng. Q. 2017, 31, 131–138. [Google Scholar] [CrossRef]
- Lima-Ramos, J.; Tufvesson, P.; Woodley, J.M. Application of environmental and economic metrics to guide the development of biocatalytic processes. Green Process. Synth. 2014, 3, 195–213. [Google Scholar] [CrossRef]
- Truppo, M.D. Biocatalysis in the pharmaceutical industry: The need for speed. ACS Med. Chem. Lett. 2017, 8, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.C.; Halder, J.M.; Nestl, B.M.; Hauer, B.; Gernaey, K.V.; Kruhne, U. Biocatalyst screening with a twist: Application of oxygen sensors integrated in microchannels for screening whole cell biocatalyst variants. Bioengineering 2018, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Lütz, S.; Giver, L.; Lalonde, J. Engineered enzymes for chemical production. Biotechnol. Bioeng. 2008, 101, 647–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tholey, A.; Heinzle, E. Methods for Biocatalyst Screening. In Tools and Applications of Biochemical Engineering Science; Schügerl, K., Zeng, A.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- Wahler, D.; Reymond, J.L. Novel methods for biocatalyst screening. Curr. Opin. Chem. Biol. 2001, 5, 152–158. [Google Scholar] [CrossRef]
- Hammerich, O.; Speiser, B. Organic Electrochemistry, Revised and Expanded, 5th ed.; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Carbonell, P.; Currin, A.; Jervis, A.J.; Rattray, N.J.W.; Swainston, N.; Yan, C.Y.; Takano, E.; Breitling, R. Bioinformatics for the synthetic biology of natural products: Integrating across the Design-Build-Test cycle. Nat. Prod. Rep. 2016, 33, 925–932. [Google Scholar] [CrossRef]
- Ellens, K.W.; Christian, N.; Singh, C.; Satagopam, V.P.; May, P.; Linster, C.L. Confronting the catalytic dark matter encoded by sequenced genomes. Nucleic Acids Res. 2017, 45, 11495–11514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popovic, A.; Hai, T.; Tchigvintsev, A.; Hajighasemi, M.; Nocek, B.; Khusnutdinova, A.N.; Brown, G.; Glinos, J.; Flick, R.; Skarina, T.; et al. Activity screening of environmental metagenomic libraries reveals novel carboxylesterase families. Sci. Rep. 2017, 7, 44103. [Google Scholar] [CrossRef] [PubMed]
- Gerlt, J.A.; Allen, K.N.; Almo, S.C.; Armstrong, R.N.; Babbitt, P.C.; Cronan, J.E.; Dunaway-Mariano, D.; Imker, H.J.; Jacobson, M.P.; Minor, W.; et al. The Enzyme Function Initiative. Biochemistry 2011, 50, 9950–9962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerlt, J.A.; Bouvier, J.T.; Davidson, D.B.; Imker, H.J.; Sadkhin, B.; Slater, D.R.; Whalen, K.L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854, 1019–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kempa, E.E.; Hollywood, K.A.; Smith, C.A.; Barran, P.E. High throughput screening of complex biological samples with mass spectrometry—From bulk measurements to single cell analysis. Analyst 2019, 144, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-L.; Wagner, J.M.; Alper, H.S. Enabling tools for high-throughput detection of metabolites: Metabolic engineering and directed evolution applications. Biotechnol. Adv. 2017, 35, 950–970. [Google Scholar] [CrossRef] [PubMed]
- Quertinmont, L.T.; Orru, R.; Lutz, S. RApid Parallel Protein EvaluatoR (RAPPER), from gene to enzyme function in one day. Chem. Commun. 2015, 51, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, X.; Yang, S.; Zhu, M.; Wang, X. Technology prospecting on enzymes: Application, marketing and engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209017. [Google Scholar] [CrossRef] [PubMed]
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Hong, S.H. Cell-free protein synthesis for producing ‘difficult-to-express’ proteins. Biochem. Eng. J. 2018, 138, 156–164. [Google Scholar] [CrossRef]
- Caschera, F.; Noireaux, V. Synthesis of 2.3 mg/mL of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie 2014, 99, 162–168. [Google Scholar] [CrossRef]
- Schwarz, D.; Junge, F.; Durst, F.; Frolich, N.; Schneider, B.; Reckel, S.; Sobhanifar, S.; Dotsch, V.; Bernhard, F. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2007, 2, 2945–2957. [Google Scholar] [CrossRef]
- Salehi, A.S.M.; Smith, M.T.; Bennett, A.M.; Williams, J.B.; Pitt, W.G.; Bundy, B.C. Cell-free protein synthesis of a cytotoxic cancer therapeutic: Onconase production and a just-add-water cell-free system. Biotechnol. J. 2016, 11, 274–281. [Google Scholar] [CrossRef]
- Sachse, R.; Dondapati, S.K.; Fenz, S.F.; Schmidt, T.; Kubick, S. Membrane protein synthesis in cell-free systems: From bio-mimetic systems to bio-membranes. FEBS Lett. 2014, 588, 2774–2781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenblum, G.; Cooperman, B.S. Engine out of the chassis: Cell-free protein synthesis and its uses. FEBS Lett. 2014, 588, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Niwa, T.; Kanamori, T.; Ueda, T.; Taguchi, H. Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc. Natl. Acad. Sci. USA 2012, 109, 8937–8942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, C.J.; Pendleton, E.D.; Sasmor, H.H.; Hicks, W.L.; Farnum, J.B.; Muto, M.; Amendt, E.M.; Schoborg, J.A.; Martin, R.W.; Clark, L.G.; et al. A cell-free expression and purification process for rapid production of protein biologics. Biotechnol. J. 2016, 11, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.W.; Des Soye, B.J.; Kwon, Y.-C.; Kay, J.; Davis, R.G.; Thomas, P.M.; Majewska, N.I.; Chen, C.X.; Marcum, R.D.; Weiss, M.G.; et al. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat. Commun. 2018, 9, 1203. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Yu, C.-H.; Liu, Y. Codon usage regulates protein structure and function by affecting translation elongation speed in Drosophila cells. Nucleic Acids Res. 2017, 45, 8484–8492. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, H.; Kanamori, T.; Shimizu, Y.; Ueda, T. A highly controllable reconstituted cell-free system-a breakthrough in protein synthesis research. Curr. Pharm. Biotechnol. 2010, 11, 267–271. [Google Scholar] [CrossRef]
- Zemella, A.; Thoring, L.; Hoffmeister, C.; Kubick, S. Cell-free protein synthesis: Pros and cons of prokaryotic and eukaryotic systems. ChemBioChem 2015, 16, 2420–2431. [Google Scholar] [CrossRef]
- Liu, W.-Q.; Zhang, L.; Chen, M.; Li, J. Cell-free protein synthesis: Recent advances in bacterial extract sources and expanded applications. Biochem. Eng. J. 2019, 141, 182–189. [Google Scholar] [CrossRef]
- Schoborg, J.A.; Hodgman, E.C.; Anderson, M.J.; Jewett, M.C. Substrate replenishment and byproduct removal improve yeast cell-free protein synthesis. Biotechnol. J. 2014, 9, 630–640. [Google Scholar] [CrossRef]
- Olliver, L.; Boyd, C.D. In vitro translation of mRNA in a rabbit reticulocyte lysate cell-free system. In The Nucleic Acid Protocols Handbook; Rapley, R., Ed.; Humana Press: Totowa, NJ, USA, 2000; pp. 885–890. [Google Scholar]
- Sawasaki, T.; Ogasawara, T.; Morishita, R.; Endo, Y. A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA 2002, 99, 14652–14657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezure, T.; Suzuki, T.; Ando, E. A cell-free protein synthesis system from insect cells. In Cell-Free Protein Synthesis: Methods and Protocols; Alexandrov, K., Johnston, W.A., Eds.; Humana Press: Totowa, NJ, USA, 2014; Volume 607, pp. 285–296. [Google Scholar]
- Brödel, A.K.; Sonnabend, A.; Kubick, S. Cell-free protein expression based on extracts from CHO cells. Biotechnol. Bioeng. 2014, 111, 25–36. [Google Scholar] [CrossRef]
- Carlson, E.D.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 2012, 30, 1185–1194. [Google Scholar] [CrossRef] [Green Version]
- Pratt, J.M. Coupled transcription-translation in prokaryotic cell-free systems. In Transcription and Translation: A Practical Approach; Hames, B.D., Higgins, S.J., Eds.; IRL Press: Oxford, UK, 1984; pp. 179–209. [Google Scholar]
- Kim, D.-M.; Swartz, J.R. Oxalate improves protein synthesis by enhancing ATP supply in a cell-free system derived from Escherichia coli. Biotechnol. Lett. 2000, 22, 1537–1542. [Google Scholar] [CrossRef]
- Jewett, M.C.; Swartz, J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004, 86, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Sitaraman, K.; Esposito, D.; Klarmann, G.; Le Grice, S.F.; Hartley, J.L.; Chatterjee, D.K. A novel cell-free protein synthesis system. J. Biotechnol. 2004, 110, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Calhoun, K.A.; Swartz, J.R. An economical method for cell-free protein synthesis using glucose and nucleoside monophosphates. Biotechnol. Prog. 2005, 21, 1146–1153. [Google Scholar] [CrossRef]
- Shin, J.; Noireaux, V. Efficient cell-free expression with the endogenous E. coli RNA polymerase and sigma factor 70. J. Biol. Eng. 2010, 4, 8. [Google Scholar] [CrossRef]
- Yang, W.C.; Patel, K.G.; Wong, H.E.; Swartz, J.R. Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol. Prog. 2012, 28, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Krinsky, N.; Kaduri, M.; Shainsky-Roitman, J.; Goldfeder, M.; Ivanir, E.; Benhar, I.; Shoham, Y.; Schroeder, A. A simple and rapid method for preparing a cell-free bacterial lysate for protein synthesis. PLoS ONE 2016, 11, e0165137. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-M.; Swartz, J.R. Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnol. Bioeng. 2001, 74, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-M.; Swartz, J.R. Prolonging cell-free protein synthesis by selective reagent additions. Biotechnol. Prog. 2000, 16, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.H. Cell-free protein synthesis energized by slowly-metabolized maltodextrin. BMC Biotechnol. 2009, 9, 58. [Google Scholar] [CrossRef]
- Ge, X.; Luo, D.; Xu, J. Cell-free protein expression under macromolecular crowding conditions. PLoS ONE 2011, 6, e28707. [Google Scholar] [CrossRef] [PubMed]
- Nevin, D.E.; Pratt, J.M. A coupled in vitro transcription-translation system for the exclusive synthesis of polypeptides expressed from the T7 promoter. FEBS Lett. 1991, 291, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, E.; Wang, Y. A modified procedure for fast purification of T7 RNA polymerase. Protein Expr. Purif. 1999, 16, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Kim, D.M.; Choi, C.Y. Rapid production of milligram quantities of proteins in a batch cell-free protein synthesis system. J. Biotechnol. 2006, 124, 373–380. [Google Scholar] [CrossRef]
- Woodrow, K.A.; Airen, I.O.; Swartz, J.R. Rapid expression of functional genomic libraries. J. Proteome Res. 2006, 5, 3288–3300. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Ivashkiv, L.; Chen, H.Z.; Zubay, G.; Cashel, M. Cell-free coupled transcription-translation system for investigation of linear DNA segments. Proc. Natl. Acad. Sci. USA 1980, 77, 7029–7033. [Google Scholar] [CrossRef]
- Michel-Reydellet, N.; Woodrow, K.; Swartz, J. Increasing PCR fragment stability and protein yields in a cell-free system with genetically modified Escherichia coli extracts. J. Mol. Microbiol. Biotechnol. 2005, 9, 26–34. [Google Scholar] [CrossRef]
- Garamella, J.; Marshall, R.; Rustad, M.; Noireaux, V. The all E. coli TX-TL toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth. Biol. 2016, 5, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Z.; Hayes, C.A.; Shin, J.; Caschera, F.; Murray, R.M.; Noireaux, V. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. JoVE 2013, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Z.; Yeung, E.; Hayes, C.A.; Noireaux, V.; Murray, R.M. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol. 2014, 3, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, A.; Hellberg, K.; Enberg, J.; Karlsson, B.G. Rational improvement of cell-free protein synthesis. New Biotechnol. 2011, 28, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Jaroentomeechai, T.; Stark, J.C.; Natarajan, A.; Glasscock, C.J.; Yates, L.E.; Hsu, K.J.; Mrksich, M.; Jewett, M.C.; DeLisa, M.P. Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat. Commun. 2018, 9, 2686. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Matsuura, T.; Sunami, T.; Kazuta, Y.; Yomo, T. In vitro evolution of alpha-hemolysin using a liposome display. Proc. Natl. Acad. Sci. USA 2013, 110, 16796–16801. [Google Scholar] [CrossRef] [PubMed]
- Damiati, S.; Kompella, U.B.; Damiati, S.A.; Kodzius, R. Microfluidic devices for drug delivery systems and drug screening. Genes 2018, 9, 103. [Google Scholar] [CrossRef]
- He, M.; Stoevesandt, O.; Taussig, M.J. In situ synthesis of protein arrays. Curr. OpiN Biotechnol. 2008, 19, 4–9. [Google Scholar] [CrossRef]
- Beppu, K.; Izri, Z.; Maeda, Y.T.; Sakamoto, R. Geometric effect for biological reactors and biological fluids. Bioengineering 2018, 5, 110. [Google Scholar] [CrossRef]
- Caschera, F.; Noireaux, V. Integration of biological parts toward the synthesis of a minimal cell. Curr. Opin. Chem. Biol. 2014, 22, 85–91. [Google Scholar] [CrossRef]
- Jiao, Y.; Liu, Y.; Luo, D.; Huck, W.T.S.; Yang, D. Microfluidic-assisted fabrication of clay microgels for cell-free protein synthesis. ACS Appl. Mater. Interfaces 2018, 10, 29308–29313. [Google Scholar] [CrossRef] [PubMed]
- Breton, M.; Amirkavei, M.; Mir, L.M. Optimization of the electroformation of giant unilamellar vesicles (GUVs) with unsaturated phospholipids. J. Membr. Biol. 2015, 248, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Elani, Y.; Law, R.V.; Ces, O. Protein synthesis in artificial cells: Using compartmentalisation for spatial organisation in vesicle bioreactors. Phys. Chem. Chem. Phys. 2015, 17, 15534–15537. [Google Scholar] [CrossRef] [PubMed]
- Los, G.V.; Encell, L.P.; McDougall, M.G.; Hartzell, D.D.; Karassina, N.; Zimprich, C.; Wood, M.G.; Learish, R.; Ohana, R.F.; Urh, M.; et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008, 3, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Heymann, M.; Bernhard, F.; Schwille, P.; Kai, L. Cell-free protein synthesis in micro compartments: Building a minimal cell from biobricks. New Biotechnol. 2017, 39, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Fenz, S.F.; Sachse, R.; Schmidt, T.; Kubick, S. Cell-free synthesis of membrane proteins: Tailored cell models out of microsomes. Biochim. Biophys. Acta 2014, 1838, 1382–1388. [Google Scholar] [CrossRef] [Green Version]
- Noireaux, V.; Libchaber, A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. USA 2004, 101, 17669–17674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.H.; Kim, D.M. Applications of cell-free protein synthesis in synthetic biology: Interfacing bio-machinery with synthetic environments. Biotechnol. J. 2013, 8, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Damiati, S.; Mhanna, R.; Kodzius, R.; Ehmoser, E.K. Cell-free approaches in synthetic biology utilizing microfluidics. Genes 2018, 9, 144. [Google Scholar] [CrossRef]
- Mei, Q.; Fredrickson, C.K.; Simon, A.; Khnouf, R.; Fan, Z.H. Cell-free protein synthesis in microfluidic array devices. Biotechnol. Prog. 2007, 23, 1305–1311. [Google Scholar] [CrossRef]
- Timm, A.C.; Shankles, P.G.; Foster, C.M.; Doktycz, M.J.; Retterer, S.T. Toward microfluidic reactors for cell-free protein synthesis at the point-of-care. Small 2016, 12, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Georgi, V.; Georgi, L.; Blechert, M.; Bergmeister, M.; Zwanzig, M.; Wustenhagen, D.A.; Bier, F.F.; Jung, E.; Kubick, S. On-chip automation of cell-free protein synthesis: New opportunities due to a novel reaction mode. Lab Chip 2016, 16, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Mazutis, L.; Baret, J.C.; Treacy, P.; Skhiri, Y.; Araghi, A.F.; Ryckelynck, M.; Taly, V.; Griffiths, A.D. Multi-step microfluidic droplet processing: Kinetic analysis of an in vitro translated enzyme. Lab Chip 2009, 9, 2902–2908. [Google Scholar] [CrossRef] [PubMed]
- Fallah-Araghi, A.; Baret, J.C.; Ryckelynck, M.; Griffiths, A.D. A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab Chip 2012, 12, 882–891. [Google Scholar] [CrossRef]
- Contreras-Llano, L.E.; Tan, C. High-throughput screening of biomolecules using cell-free gene expression systems. Synth. Biol. 2018, 3. [Google Scholar] [CrossRef]
- He, M.; Taussig, M.J. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method). Nucleic Acids Res. 2001, 29, e73. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, N.; Hainsworth, E.; Bhullar, B.; Eisenstein, S.; Rosen, B.; Lau, A.Y.; Walter, J.C.; LaBaer, J. Self-assembling protein microarrays. Science 2004, 305, 86–90. [Google Scholar] [CrossRef]
- Song, L.; Wallstrom, G.; Yu, X.; Hopper, M.; Van Duine, J.; Steel, J.; Park, J.; Wiktor, P.; Kahn, P.; Brunner, A.; et al. Identification of antibody targets for tuberculosis serology using high-density nucleic acid programmable protein arrays. Mol. Cell. Proteom. 2017, 16, 277–289. [Google Scholar] [CrossRef]
- He, M.; Stoevesandt, O.; Palmer, E.A.; Khan, F.; Ericsson, O.; Taussig, M.J. Printing protein arrays from DNA arrays. Nat. Methods 2008, 5, 175. [Google Scholar] [CrossRef]
- Schmidt, R.; Cook, E.A.; Kastelic, D.; Taussig, M.J.; Stoevesandt, O. Optimised ‘on demand’ protein arraying from DNA by cell free expression with the ‘DNA to Protein Array’ (DAPA) technology. J. Proteom. 2013, 88, 141–148. [Google Scholar] [CrossRef]
- Catherine, C.; Lee, K.H.; Oh, S.J.; Kim, D.M. Cell-free platforms for flexible expression and screening of enzymes. Biotechnol. Adv. 2013, 31, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhao, J.; Lian, J.; Xu, Z. Cell-free protein synthesis enabled rapid prototyping for metabolic engineering and synthetic biology. Synth. Syst. Biotechnol. 2018, 3, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Murthy, T.V.; Wu, W.; Qiu, Q.Q.; Shi, Z.; LaBaer, J.; Brizuela, L. Bacterial cell-free system for high-throughput protein expression and a comparative analysis of Escherichia coli cell-free and whole cell expression systems. Protein Expr. Purif. 2004, 36, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Kim, K.S.; Kang, T.J.; Choi, J.H.; Song, J.J.; Choi, Y.H.; Kim, B.G.; Kim, D.M. Implementing bacterial acid resistance into cell-free protein synthesis for buffer-free expression and screening of enzymes. Biotechnol. Bioeng. 2015, 112, 2630–2635. [Google Scholar] [CrossRef]
- Quertinmont, L.T.; Lutz, S. Cell-free protein engineering of Old Yellow Enzyme 1 from Saccharomyces pastorianus. Tetrahedron 2016, 72, 7282–7287. [Google Scholar] [CrossRef]
- Kelwick, R.; Ricci, L.; Webb, A.J.; Freemont, P.S.; Bell, D.; Chee, S.M. Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Synth. Biol. 2018, 3, 1–44. [Google Scholar] [CrossRef]
- Dudley, Q.M.; Karim, A.S.; Jewett, M.C. Cell-free metabolic engineering: Biomanufacturing beyond the cell. Biotechnol. J. 2015, 10, 69–82. [Google Scholar] [CrossRef]
- Morgado, G.; Gerngross, D.; Roberts, T.M.; Panke, S. Synthetic biology for cell-free biosynthesis: Fundamentals of designing novel in vitro multi-enzyme reaction networks. In Synthetic Biology—Metabolic Engineering; Zhao, H., Zeng, A.-P., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 117–146. [Google Scholar]
- Dudley, Q.M.; Anderson, K.C.; Jewett, M.C. Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol. 2016, 5, 1578–1588. [Google Scholar] [CrossRef]
- Karim, A.S.; Jewett, M.C. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 2016, 36, 116–126. [Google Scholar] [CrossRef] [Green Version]
- Casini, A.; Chang, F.Y.; Eluere, R.; King, A.M.; Young, E.M.; Dudley, Q.M.; Karim, A.; Pratt, K.; Bristol, C.; Forget, A.; et al. A pressure test to make 10 molecules in 90 days: External evaluation of methods to engineer biology. J. Am. Chem. Soc. 2018, 140, 4302–4316. [Google Scholar] [CrossRef]
- Fujiwara, K.; Doi, N. Biochemical preparation of cell extract for cell-free protein synthesis without physical disruption. PLoS ONE 2016, 11, e0154614. [Google Scholar] [CrossRef] [PubMed]
- Zawada, J.F.; Yin, G.; Steiner, A.R.; Yang, J.; Naresh, A.; Roy, S.M.; Gold, D.S.; Heinsohn, H.G.; Murray, C.J. Microscale to manufacturing scale-up of cell-free cytokine production—A new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011, 108, 1570–1578. [Google Scholar] [CrossRef] [PubMed]
- Swartz, J.R. Expanding biological applications using cell-free metabolic engineering: An overview. Metab. Eng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Saurabh, S.; Bruchez, M.P.; Schwartz, R.; Leduc, P. Molecular crowding shapes gene expression in synthetic cellular nanosystems. Nat. Nanotechnol. 2013, 8, 602–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, B.R.; Jamil, O.K.; Jewett, M.C. Implications of macromolecular crowding and reducing conditions for in vitro ribosome construction. Nucleic Acids Res. 2015, 43, 4774–4784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, N.-N.; Vibhute, M.A.; Zheng, L.; Zhao, H.; Yelleswarapu, M.; Huck, W.T.S. Macromolecularly crowded protocells from reversibly shrinking monodisperse liposomes. J. Am. Chem. Soc. 2018, 140, 7399–7402. [Google Scholar] [CrossRef]
- Okano, T.; Matsuura, T.; Suzuki, H.; Yomo, T. Cell-free protein synthesis in a microchamber revealed the presence of an optimum compartment volume for high-order reactions. ACS Synth. Biol. 2014, 3, 347–352. [Google Scholar] [CrossRef]
- Sakamoto, R.; Noireaux, V.; Maeda, Y.T. Anomalous scaling of gene expression in confined cell-free reactions. Sci. Rep. 2018, 8, 7364. [Google Scholar] [CrossRef]
- Niess, A.; Failmezger, J.; Kuschel, M.; Siemann-Herzberg, M.; Takors, R. Experimentally validated model enables debottlenecking of in vitro protein synthesis and identifies a control shift under in vivo conditions. ACS Synth. Biol. 2017, 6, 1913–1921. [Google Scholar] [CrossRef]
- Hansen, M.M.; Ventosa Rosquelles, M.; Yelleswarapu, M.; Maas, R.J.; van Vugt-Jonker, A.J.; Heus, H.A.; Huck, W.T. Protein synthesis in coupled and uncoupled cell-free prokaryotic gene expression systems. ACS Synth. Biol. 2016, 5, 1433–1440. [Google Scholar] [CrossRef]
- Hurst, G.B.; Asano, K.G.; Doktycz, C.J.; Consoli, E.J.; Doktycz, W.L.; Foster, C.M.; Morrell-Falvey, J.L.; Standaert, R.F.; Doktycz, M.J. Proteomics-based tools for evaluation of cell-free protein synthesis. Anal. Chem. 2017, 89, 11443–11451. [Google Scholar] [CrossRef] [PubMed]
- Foshag, D.; Henrich, E.; Hiller, E.; Schafer, M.; Kerger, C.; Burger-Kentischer, A.; Diaz-Moreno, I.; Garcia-Maurino, S.M.; Dotsch, V.; Rupp, S.; et al. The E. coli S30 lysate proteome: A prototype for cell-free protein production. New Biotechnol. 2018, 40, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Vilkhovoy, M.; Horvath, N.; Shih, C.H.; Wayman, J.A.; Calhoun, K.; Swartz, J.; Varner, J.D. Sequence specific modeling of E. coli cell-free protein synthesis. ACS Synth. Biol. 2018, 7, 1844–1857. [Google Scholar] [CrossRef] [PubMed]
- Gagoski, D.; Shi, Z.; Nielsen, L.K.; Vickers, C.E.; Mahler, S.; Speight, R.; Johnston, W.A.; Alexandrov, K. Cell-free pipeline for discovery of thermotolerant xylanases and endo-1,4-β-glucanases. J. Biotechnol. 2017, 259, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Borkowski, O.; Bricio, C.; Murgiano, M.; Rothschild-Mancinelli, B.; Stan, G.B.; Ellis, T. Cell-free prediction of protein expression costs for growing cells. Nat. Commun. 2018, 9, 1457. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.; Maxwell, C.S.; Collins, S.P.; Jacobsen, T.; Luo, M.L.; Begemann, M.B.; Gray, B.N.; January, E.; Singer, A.; He, Y.H.; et al. Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell 2018, 69, 146–157. [Google Scholar] [CrossRef] [PubMed]




| Protein | CAT | CAT | CAT | eGFP | CAT | deGFP | sfGFP | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Yield [µg mL−1] | - | 194 | 700 | - | 800 | - | 639 | 700 | 2300 | 150 | |
| Purpose | Component | Pratt, 1984 [49] | Kim, 2000 [50] | Jewett, 2004 [51] | Sitaraman, 2004 [52] | Calhoun, 2005 [53] | Ohashi, 2010 [40] | Shin, 2010 [54] | Yang, 2012 [55] | Caschera, 2014 [31] | Krinsky, 2016 [56] |
| Substrates for protein synthesis | Amino acids | X | X | X | X | X | X | X | X | X | X |
| Energy source; substrate | ATP | X | X | X | X | - | X | X | X | X | X |
| Substrates for transcription | GTP, CTP, UTP | X | X | X | X | - | X | X | - | X | X |
| Reduction of reaction costs | Nucleoside monophosphates | - | - | - | - | X | - | - | X | - | - |
| Supply of amino acids | tRNA | X | X | X | X | X | X | X | X | X | - |
| Buffer | HEPES | - | X | - | X | X | X | X | - | X | X |
| Stabilizes polymerases | DTT | X | X | - | X | - | X | X | - | X | - |
| Increased yields | cAMP | X | X | - | X | - | X | X | - | X | - |
| Formation of initiator formyl-methionine | Folinic acid | X | X | X | X | X | X | X | X | X | - |
| Viscosity; stability of mRNA; crowding effects | Polyethylene glycol | X | X | - | X | - | - | X | X | X | X |
| Energy regeneration; phosphorylation of nucleoside monophosphates | Creatine phosphate | - | - | - | - | - | X | - | - | - | - |
| Phosphoenolpyruvate | X | X | - | - | X | - | - | X | - | - | |
| Pyruvate | - | - | X | - | - | - | - | - | - | - | |
| 3-Phosphoglyceric acid | - | - | - | X | - | - | X | - | X | X | |
| Recycling of inorganic phosphate | Maltose | - | - | - | - | - | - | - | - | X | - |
| Regeneration of ATP from pyruvate | NAD | - | X | X | - | X | - | X | X | X | - |
| Coenzyme A | - | X | X | - | X | - | X | X | X | - | |
| Stabilization of nucleic acids; stimulate polymerase activity | Spermidine | - | - | X | - | X | X | X | X | X | - |
| Putrescine | - | - | X | - | X | - | - | X | - | - | |
| Inhibitor of PEP synthetase | Oxalate | - | X | X | - | X | - | - | - | - | - |
| Kations and anions | Mg2+, K+, NH4+, acetate, glutamate | X | X | X | X | X | X | X | X | X | X |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rolf, J.; Rosenthal, K.; Lütz, S. Application of Cell-Free Protein Synthesis for Faster Biocatalyst Development. Catalysts 2019, 9, 190. https://doi.org/10.3390/catal9020190
Rolf J, Rosenthal K, Lütz S. Application of Cell-Free Protein Synthesis for Faster Biocatalyst Development. Catalysts. 2019; 9(2):190. https://doi.org/10.3390/catal9020190
Chicago/Turabian StyleRolf, Jascha, Katrin Rosenthal, and Stephan Lütz. 2019. "Application of Cell-Free Protein Synthesis for Faster Biocatalyst Development" Catalysts 9, no. 2: 190. https://doi.org/10.3390/catal9020190
APA StyleRolf, J., Rosenthal, K., & Lütz, S. (2019). Application of Cell-Free Protein Synthesis for Faster Biocatalyst Development. Catalysts, 9(2), 190. https://doi.org/10.3390/catal9020190