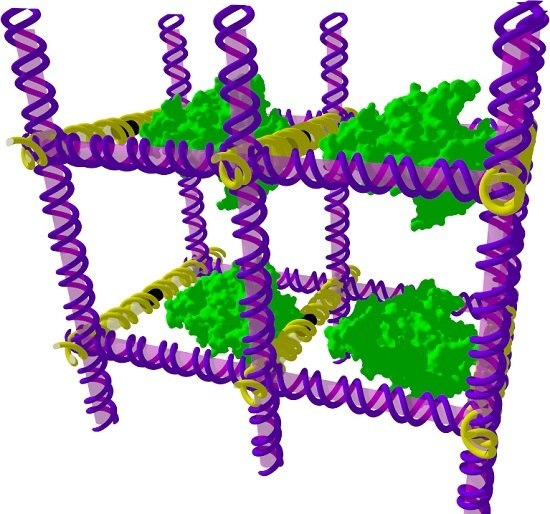
3D DNA Crystals and Nanotechnology
Abstract
:1. Introduction
2. Tensegrity Triangle Crystals
3. DNA 13-Mer Crystals with Non-Canonical Base Pairs
4. Lessons for DNA Crystal Design
5. DNA Crystal Applications
6. Future Directions and Applications
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Franklin, R.E.; Gosling, R.G. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C.; Sussman, J.L.; Berman, H.M.; Kim, S.H. Nucleic Acid Conformation: Crystal Structure of a Naturally Occurring Dinucleoside Phosphate (UpA). Nature 1971, 233, 90–92. [Google Scholar] [CrossRef]
- Caruthers, M.H. Gene synthesis machines: DNA chemistry and its uses. Science 1985, 230, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.H.-J.; Quigley, G.J.; Kolpak, F.J.; Crawford, J.L.; van Boom, J.H.; van der Marel, G.; Rich, A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 1979, 282, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [PubMed]
- Brown, T.; Hunter, W.N.; Kneale, G.; Kennard, O. Molecular structure of the G.A base pair in DNA and its implications for the mechanism of transversion mutations. Proc. Natl. Acad. Sci. USA 1986, 83, 2402–2406. [Google Scholar] [CrossRef] [PubMed]
- Sines, C.C.; McFail-Isom, L.; Howerton, S.B.; VanDerveer, D.; Williams, L.D. Cations Mediate B-DNA Conformational Heterogeneity. J. Am. Chem. Soc. 2000, 122, 11048–11056. [Google Scholar] [CrossRef]
- Hays, F.A.; Teegarden, A.; Jones, Z.J.R.; Harms, M.; Raup, D.; Watson, J.; Cavaliere, E.; Ho, P.S. How sequence defines structure: A crystallographic map of DNA structure and conformation. Proc. Natl. Acad. Sci. USA 2004, 102, 7157–7162. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Geiger, J.H.; Hahn, S.; Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 1993, 365, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [PubMed]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Robinson, B.H.; Seeman, N.C. The design of a biochip: A self-assembling molecular-scale memory device. Protein Eng. 1987, 1, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Paukstelis, P.J. Three-dimensional DNA crystals as molecular sieves. J. Am. Chem. Soc. 2006, 128, 6794–6795. [Google Scholar] [CrossRef] [PubMed]
- Geng, C.; Paukstelis, P.J. DNA Crystals as Vehicles for Biocatalysis. J. Am. Chem. Soc. 2014, 136, 7817–7820. [Google Scholar] [CrossRef] [PubMed]
- Inokuma, Y.; Yoshioka, S.; Ariyoshi, J.; Arai, T.; Hitora, Y.; Takada, K.; Matsunaga, S.; Rissanen, K.; Fujita, M. X-ray analysis on the nanogram to microgram scale using porous complexes. Nature 2013, 495, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Moon, S.-Y.; Guelta, M.A.; Harvey, S.P.; Hupp, J.T.; Farha, O.K. Encapsulation of a Nerve Agent Detoxifying Enzyme by a Mesoporous Zirconium Metal-Organic Framework Engenders Thermal and Long-Term Stability. J. Am. Chem. Soc. 2016, 138, 8052–8055. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Dewan, J.C.; Seeman, N.C. A DNA decamer with a sticky end: The crystal structure of d-CGACGATCGT. J. Mol. Biol. 1997, 267, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.-S.; Wu, H.-M.; Crothers, D.M. DNA bending at adenine-thymine tracts. Nature 1986, 320, 501–506. [Google Scholar] [CrossRef] [PubMed]
- DiGabriele, A.D.; Steitz, T.A. A DNA dodecamer containing an adenine tract crystallizes in a unique lattice and exhibits a new bend. J. Mol. Biol. 1993, 231, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.H.; Wartell, R.M. The thermal stability of DNA fragments with tandem mismatches at a d(CXYG).d(CY’X’G) site. Nucleic Acids Res. 1996, 24, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, P.; Ohmichi, T.; Sugimoto, N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002, 526, 77–81. [Google Scholar] [CrossRef]
- Mendoza, O.; Elezgaray, J.; Mergny, J.-L. Kinetics of quadruplex to duplex conversion. Biochimie 2015, 118, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Thirugnanasambandam, A.; Karthik, S.; Artheswari, G.; Gautham, N. DNA polymorphism in crystals: Three stable conformations for the decadeoxynucleotide d(GCATGCATGC). Acta Crystallogr. Sect. Struct. Biol. 2016, 72, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C.; Rosenberg, J.M.; Rich, A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. USA 1976, 73, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Urbina, G.; Sathe, G.M.; Liu, W.C.; Gillen, M.F.; Duck, P.D.; Bender, R.; Ogilvie, K.K. Automated synthesis of gene fragments. Science 1981, 214, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Structural DNA Nanotechnology; Cambridge University Press: Cambridge, UK, 2015. [Google Scholar]
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-Dimensional Structures Self-Assembled from DNA Bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Marchi, A.N.; Saaem, I.; Vogen, B.N.; Brown, S.; LaBean, T.H. Toward Larger DNA Origami. Nano Lett. 2014, 14, 5740–5747. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, P.J. Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 1988, 17, 265–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Seeman, N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991, 350, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Seeman, N.C. Symmetric Holliday junction crossover isomers. J. Mol. Biol. 1994, 238, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.J.; Seeman, N.C. DNA double-crossover molecules. Biochemistry 1993, 32, 3211–3220. [Google Scholar] [CrossRef] [PubMed]
- LaBean, T.H.; Yan, H.; Kopatsch, J.; Liu, F.; Winfree, E.; Reif, J.H.; Seeman, N.C. Construction, Analysis, Ligation, and Self-Assembly of DNA Triple Crossover Complexes. J. Am. Chem. Soc. 2000, 122, 1848–1860. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Seeman, N.C. Direct evidence for Holliday junction crossover isomerization. Biochemistry 1997, 36, 4240–4247. [Google Scholar] [CrossRef] [PubMed]
- Sa-Ardyen, P.; Vologodskii, A.V.; Seeman, N.C. The flexibility of DNA double crossover molecules. Biophys. J. 2003, 84, 3829–3837. [Google Scholar] [CrossRef]
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, F.; Liao, S.; Kopatsch, J.; Wang, T.; Mao, C.; Seeman, N.C. Six-helix bundles designed from DNA. Nano Lett. 2005, 5, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; LaBean, T.H.; Reif, J.H.; Seeman, N.C. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 2000, 407, 493–496. [Google Scholar] [PubMed]
- Mao, C.; Sun, W.; Shen, Z.; Seeman, N.C. A nanomechanical device based on the B-Z transition of DNA. Nature 1999, 397, 144–146. [Google Scholar] [PubMed]
- Yan, H.; Zhang, X.; Shen, Z.; Seeman, N.C. A robust DNA mechanical device controlled by hybridization topology. Nature 2002, 415, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Seeman, N.C. Circuits and programmable self-assembling DNA structures. Proc. Natl. Acad. Sci. USA 2002, 99, 12577–12582. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Ong, L.L.; Sun, W.; Song, J.; Dong, M.; Shih, W.M.; Yin, P. DNA brick crystals with prescribed depths. Nat. Chem. 2014, 6, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lytton-Jean, A.K.R.; Lee, B.; Weigand, S.; Schatz, G.C.; Mirkin, C.A. DNA-programmable nanoparticle crystallization. Nature 2008, 451, 553–556. [Google Scholar] [CrossRef] [PubMed]
- Nykypanchuk, D.; Maye, M.M.; van der Lelie, D.; Gang, O. DNA-guided crystallization of colloidal nanoparticles. Nature 2008, 451, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Um, S.H.; Lee, J.B.; Park, N.; Kwon, S.Y.; Umbach, C.C.; Luo, D. Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006, 5, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, A.; Frenkel, D. Numerical Evidence for Nucleated Self-Assembly of DNA Brick Structures. Phys. Rev. Lett. 2014, 112, 238103. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, B.C.; Westbrook, J.; Ghosh, S.; Petrov, A.I.; Sweeney, B.; Zirbel, C.L.; Leontis, N.B.; Berman, H.M. The Nucleic Acid Database: New features and capabilities. Nucleic Acids Res. 2014, 42, D114–D122. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, M.; Deng, Z.; Walulu, R.; Mao, C. Tensegrity: Construction of Rigid DNA Triangles with Flexible Four-Arm DNA Junctions. J. Am. Chem. Soc. 2004, 126, 2324–2325. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Birktoft, J.J.; Chen, Y.; Wang, T.; Sha, R.; Constantinou, P.E.; Ginell, S.L.; Mao, C.; Seeman, N.C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 2009, 461, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Sha, R.; Birktoft, J.; Zheng, J.; Mao, C.; Seeman, N.C. A DNA Crystal Designed to Contain Two Molecules per Asymmetric Unit. J. Am. Chem. Soc. 2010, 132, 15471–15473. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhong, H.; Wang, R.; Seeman, N.C. Crystalline Two-Dimensional DNA Origami Arrays. Angew. Chem. Int. Ed. Engl. 2011, 50, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Paukstelis, P.J.; Nowakowski, J.; Birktoft, J.J.; Seeman, N.C. Crystal Structure of a Continuous Three-Dimensional DNA Lattice. Chem. Biol. 2004, 11, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Nowakowski, J.; Shim, P.J.; Prasad, G.S.; Stout, C.D.; Joyce, G.F. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10–23 DNA enzyme. Nat. Struct. Mol. Biol. 1999, 6, 151–156. [Google Scholar]
- Chakraborty, S.; Sharma, S.; Maiti, P.K.; Krishnan, Y. The poly dA helix: A new structural motif for high performance DNA-based molecular switches. Nucleic Acids Res. 2009, 37, 2810–2817. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Chakraborty, K.; Krishnan, Y. Tunable, colorimetric DNA-based pH sensors mediated by A-motif formation. Chem. Commun. 2012, 48, 2513–2515. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.; Nizak, C.; Surana, S.; Halder, S.; Krishnan, Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 2013, 8, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Marsh, T.C.; Henderson, E. G-Wires: Self-Assembly of a Telomeric Oligonucleotide, d(GGGGTTGGGG), into Large Superstructures. Biochemistry 1994, 33, 10718–10724. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yang, Z.; Liu, D. DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 2014, 47, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Kettani, A.; Bouaziz, S.; Skripkin, E.; Majumdar, A.; Wang, W.; Jones, R.A.; Patel, D.J. Interlocked mismatch-aligned arrowhead DNA motifs. Struct. Lond. Engl. 1999, 7, 803–815. [Google Scholar] [CrossRef]
- Robinson, H.; van der Marel, G.A.; van Boom, J.H.; Wang, A.H. Unusual DNA conformation at low pH revealed by NMR: Parallel-stranded DNA duplex with homo base pairs. Biochemistry 1992, 31, 10510–10517. [Google Scholar] [CrossRef] [PubMed]
- Robinson, H.; Wang, A.H. 5’-CGA sequence is a strong motif for homo base-paired parallel-stranded DNA duplex as revealed by NMR analysis. Proc. Natl. Acad. Sci. USA 1993, 90, 5224–5228. [Google Scholar] [CrossRef] [PubMed]
- Saoji, M.; Zhang, D.; Paukstelis, P.J. Probing the role of sequence in the assembly of three-dimensional DNA crystals. Biopolymers 2015, 103, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Seeman, N.C.; Paukstelis, P.J. Surface AFM of a 3D DNA crystal. University of Maryland: College Park, MD, USA, Unpublished work. 2010. [Google Scholar]
- Berger, I.; Kang, C.; Sinha, N.; Wolters, M.; Rich, A. A Highly Efficient 24-Condition Matrix for the Crystallization of Nucleic Acid Fragments. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Viladoms, J.; Parkinson, G.N. HELIX: A new modular nucleic acid crystallization screen. J. Appl. Crystallogr. 2014, 47, 948–955. [Google Scholar] [CrossRef]
- Ohayon, Y.P.; Chandrasekaran, A.R.; Hernandez, C.; Jong, M.A.; Abdallah, H.O.; Fisher, N.; Mohsen, M.; Shah, C.; Demirel, E.; Tan, A.; et al. Designing Higher Resolution Self-Assembled 3D DNA Crystals via Strand Terminus Modifications. New York University: New York, NY, USA, Unpublished work. 2016. [Google Scholar]
- Sha, R.; Birktoft, J.J.; Nguyen, N.; Chandrasekaran, A.R.; Zheng, J.; Zhao, X.; Mao, C.; Seeman, N.C. Self-Assembled DNA Crystals: The Impact on Resolution of 5′-Phosphates and the DNA Source. Nano Lett. 2013, 13, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Boistelle, R.; Astier, J.P. Crystallization mechanisms in solution. J. Cryst. Growth 1988, 90, 14–30. [Google Scholar] [CrossRef]
- Kantardjieff, K.A.; Rupp, B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein–nucleic acid complex crystals. Protein Sci. 2003, 12, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Song, C.; Nangreave, J.; Liu, X.; Lin, L.; Qiu, D.; Wang, Z.-G.; Zou, G.; Liang, X.; Yan, H.; et al. DNA Origami as a Carrier for Circumvention of Drug Resistance. J. Am. Chem. Soc. 2012, 134, 13396–13403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA Origami as an In Vivo Drug Delivery Vehicle for Cancer Therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, A.L.; Mrsny, R.J. Formulation and delivery issues for monoclonal antibody therapeutics. Adv. Drug Deliv. Rev. 2006, 58, 686–706. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Qian, H.; Tay, C.Y.; Leong, D.T. Cellular processing and destinies of artificial DNA nanostructures. Chem. Soc. Rev. 2016, 45, 4199–4225. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.S.; Yin, H.; Erben, C.M.; Wood, M.J.A.; Turberfield, A.J. DNA Cage Delivery to Mammalian Cells. ACS Nano 2011, 5, 5427–5432. [Google Scholar] [CrossRef] [PubMed]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef] [PubMed]
- Hahn, J.; Wickham, S.F.J.; Shih, W.M.; Perrault, S.D. Addressing the Instability of DNA Nanostructures in Tissue Culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Photo-Cross-Linking-Assisted Thermal Stability of DNA Origami Structures and Its Application for Higher-Temperature Self-Assembly. J. Am. Chem. Soc. 2011, 133, 14488–14491. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, V.; Oberleitner, B.; Sobotta, J.; Nickels, P.; Grossi, G.; Kempter, S.; Frischmuth, T.; Liedl, T.; Manetto, A. One-Step Formation of “Chain-Armor”-Stabilized DNA Nanostructures. Angew. Chem. Int. Ed. Engl. 2015, 54, 7795–7798. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chandrasekaran, A.R.; Li, Q.; Li, X.; Sha, R.; Seeman, N.C.; Mao, C. Post-Assembly Stabilization of Rationally Designed DNA Crystals. Angew. Chem. Int. Ed. 2015, 54, 9936–9939. [Google Scholar] [CrossRef] [PubMed]
- Rusling, D.A.; Chandrasekaran, A.R.; Ohayon, Y.P.; Brown, T.; Fox, K.R.; Sha, R.; Mao, C.; Seeman, N.C. Functionalizing Designer DNA Crystals with a Triple-Helical Veneer. Angew. Chem. Int. Ed. Engl. 2014, 53, 3979–3982. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.O.; Ohayon, Y.P.; Chandrasekaran, A.R.; Sha, R.; Fox, K.R.; Brown, T.; Rusling, D.A.; Mao, C.; Seeman, N.C. Stabilisation of self-assembled DNA crystals by triplex-directed photo-cross-linking. Chem. Commun. 2016, 52, 8014–8017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Paukstelis, P.J. Enhancing DNA Crystal Durability through Chemical Crosslinking. ChemBioChem 2016, 17, 1163–1170. [Google Scholar] [CrossRef] [PubMed]
- Boerneke, M.A.; Dibrov, S.M.; Hermann, T. Crystal-Structure-Guided Design of Self-Assembling RNA Nanotriangles. Angew. Chem. Int. Ed. Engl. 2016, 55, 4097–4100. [Google Scholar] [CrossRef] [PubMed]








© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paukstelis, P.J.; Seeman, N.C. 3D DNA Crystals and Nanotechnology. Crystals 2016, 6, 97. https://doi.org/10.3390/cryst6080097
Paukstelis PJ, Seeman NC. 3D DNA Crystals and Nanotechnology. Crystals. 2016; 6(8):97. https://doi.org/10.3390/cryst6080097
Chicago/Turabian StylePaukstelis, Paul J., and Nadrian C. Seeman. 2016. "3D DNA Crystals and Nanotechnology" Crystals 6, no. 8: 97. https://doi.org/10.3390/cryst6080097
APA StylePaukstelis, P. J., & Seeman, N. C. (2016). 3D DNA Crystals and Nanotechnology. Crystals, 6(8), 97. https://doi.org/10.3390/cryst6080097