Mechanochemical Synthesis and Crystal Structure of the Lidocaine-Phloroglucinol Hydrate 1:1:1 Complex †
Abstract
:1. Introduction
2. Materials and Methods
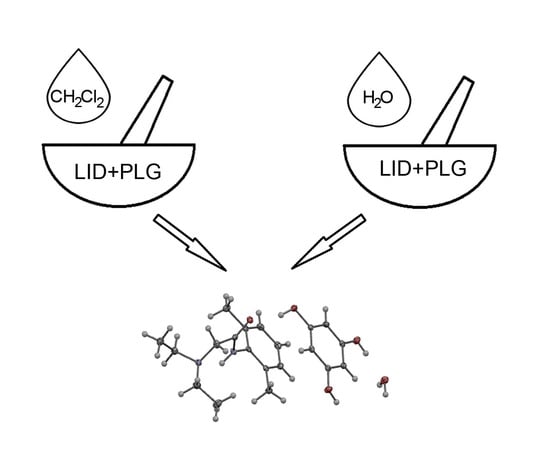
2.1. Mechanochemical Synthesis and Crystallization
2.2. IR Spectroscopy
2.3. X-Ray Diffraction
3. Results and Discussion
3.1. Infrared Spectroscopy
3.2. X-Ray Diffraction
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Allen, L.V.; Popovich, N.G.; Ansel, H.C. Ansel’s Pharmaceutical Dosage Forms and Drug Delivery Systems, 9th ed.; Lippincott & Wilkins: Baltimore, MD, USA, 2011; pp. 90–92. [Google Scholar]
- Healy, A.M.; Worku, Z.A.; Kumar, D.; Madi, A.M. Pharmaceutical solvates, hydrates and amorphous forms: A special emphasis on cocrystals. Adv. Drug Deliv. Rev. 2017, 117, 25–46. [Google Scholar] [CrossRef] [PubMed]
- Umeda, Y.; Fukami, T.; Furuishi, T.; Suzuki, T.; Makimura, M.; Tomono, K. Molecular Complex Consisting of Two Typical External Medicines: Intermolecular Interaction between Indomethacin and Lidocaine. Chem. Pharm. Bull. 2007, 55, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Steed, J.W.; Atwood, J.L. Supramolecular Chemistry, 2nd ed.; John Wiley & Sons Ltd: Wiltshire, UK, 2009; pp. 2–9. [Google Scholar]
- Ferreira, S.M.; Campos, K.D. The analgesic effect of intravenous lidocaine in the treatment of chronic pain: A literature review. Rev. Bras. Reumatol. 2014, 54, 386–392. [Google Scholar]
- Badawi, H.M.; Förner, W.; Ali, S.A. The molecular structure and vibrational, 1H and 13C NMR spectra of lidocaine hydrochloride monohydrate. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 152, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Rajendiran, N.; Mohandoss, T.; Saravanan, J. Guest: Host interactions of lidocaine and prilocaine with natural cyclodextrins: Spectral and molecular modeling studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 132, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Umeda, Y.; Nagase, H.; Makimura, M.; Tomono, K.; Shiro, M.; Ueda, H. Crystal Structure of a 2:1 Complex of Indomethacin and Lidocaine. Anal. Sci. 2007, 23, x15–x16. [Google Scholar] [CrossRef]
- Corvis, Y.; Négrier, P.; Lazerges, M.; Massip, S.; Léger, J.M.; Espeau, P. Lidocaine/L-Menthol Binary System: Cocrystallization versus Solid-State Immiscibility. J. Phys. Chem. B 2010, 114, 5420–5426. [Google Scholar] [CrossRef] [PubMed]
- Lebedyeva, I.O.; Oliferenko, A.A.; Oliferenko, P.V.; Hromas, R.A.; Neubert, J.K.; Caudle, R.M.; Wickersham, J.; Castleman, W.L.; Altschuler, G.I.; Ostrov, D.A.; et al. Ionic conjugates of lidocaine and sweeteners as better tasting local anesthetics for dentistry. J. Mater. Chem. B 2015, 3, 8492–8498. [Google Scholar] [CrossRef]
- Berton, P.; Di Bona, K.R.; Yancey, D.; Rizvi, S.A.A.; Gray, M.; Gurau, G.; Shamshina, J.L.; Rasco, J.F.; Rogers, R.D. Transdermal Bioavailability in Rats of Lidocaine in the Forms of Ionic Liquids, Salts, and Deep Eutectic. ACS Med. Chem. Lett. 2017, 8, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Enkelmann, V.; Brunklaus, G. O-H ··· N Heterosynthon: A Robust Supramolecular Unit for Crystal Engineering. Cryst. Growth Des. 2009, 9, 2354–2362. [Google Scholar] [CrossRef]
- Mukherjee, A.; Grobelny, P.; Thakur, T.S.; Desiraju, G.R. Polymorphs, Pseudopolymorphs, and Co-Crystals of Orcinol: Exploring the Structural Landscape with High Throughput Crystallography. Cryst. Growth Des. 2011, 11, 2637–2653. [Google Scholar] [CrossRef]
- Sanphui, P.; Goud, N.R.; Khandavilli, U.B.R.; Nangia, A. Fast Dissolving Curcumin Cocrystals. Cryst. Growth Des. 2011, 11, 4135–4145. [Google Scholar] [CrossRef]
- Karki, S.; Friscic, T.; Fabian, L.; Jones, W. New solid forms of artemisinin obtained through cocrystallisation. CrystEngComm 2010, 12, 4038–4041. [Google Scholar] [CrossRef]
- Swapna, B.; Maddileti, D.; Nangia, A. Cocrystals of the Tuberculosis Drug Isoniazid: Polymorphism, Isostructurality, and Stability. Cryst. Growth Des. 2014, 14, 5991–6005. [Google Scholar] [CrossRef]
- Tacaks, L. The historical development of mechanochemistry. Chem. Soc. Rev. 2013, 42, 7649–7659. [Google Scholar] [CrossRef] [PubMed]
- Delori, A.; Friscic, T.; Jones, W. The role of mechanochemistry and supramolecular design in the development of pharmaceutical materials. CrystEngComm 2012, 14, 2350–2362. [Google Scholar] [CrossRef]
- Tan, D.; Loots, L.; Friscic, T. Towards medicinal mechanochemistry: Evolution of milling from pharmaceutical solid form screening to the synthesis of active pharmaceutical ingredients (APIs). Chem. Commun. 2016, 52, 7760–7781. [Google Scholar] [CrossRef] [PubMed]
- Do, J.L.; Friscic, T. Mechanochemistry: A Force of Synthesis. ACS Cent. Sci. 2017, 3, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Hanson, A.W.; Banner, D.W. 2-Diethylamino-2′,6′-acetoxylidide (lidocaine). Acta Crystallogr. B Struct. Crystallogr. Cryst. Chem. 1974, 30, 2486–2488. [Google Scholar] [CrossRef]
- Bruker, A.X.S. APEX2, SAINT and SADABS; Bruker AXS Inc.: Madison, WI, USA, 2008. [Google Scholar]
- Blessing, R.H. An empirical correction for absorption anisotropy. Acta Crystallogr. Sect. A Found. Crystallogr. 1995, 51, 33–38. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Florey, K. Analytical Profiles of Drug Substances; Academic Press: Orlando, FL, USA, 1986; Volume 15, pp. 761–779. [Google Scholar]
- Braun, D.E.; Tocher, D.A.; Price, S.L.; Griesser, U.J. The Complexity of Hydration of Phloroglucinol: A Comprehensive Structural and Thermodynamic Characterization. J. Phys. Chem. B 2012, 116, 3961–3972. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-Balderas, M.M.; Delgado-Alfaro, R.A.; Martínez-Martínez, F.J.; Ortegón-Reyna, D.; Bernabé-Pineda, M.; Zúñiga-Lemus, O.; González-González, J.S. Synthesis, Molecular Structure of Diethyl Phenylenebis(Methylene)Dicarbamates and FTIR Spectroscopy Molecular Recognition Study with Benzenediols. J. Braz. Chem. Soc. 2015, 26, 396–402. [Google Scholar] [CrossRef]
- González-González, J.S.; Martínez-Martínez, F.J.; García-Báez, E.V.; Cruz, A.; Morín-Sánchez, L.M.; Rojas-Lima, S.; Padilla-Martínez, I.I. Molecular Complexes of Diethyl N,N′-1,3-Phenyldioxalamate and Resorcinols: Conformational Switching through Intramolecular Three-Centered Hydrogen-Bonding. Cryst. Growth Des. 2014, 14, 628–642. [Google Scholar] [CrossRef]
- CCDC Access Structures. Available online: https://www.ccdc.cam.ac.uk/structures/?ccdc-check=1afe22a817ae3814a38887d4ad47aa67 (accessed on January 20th).







| LID-PLG(Cryst) | |
|---|---|
| CCDC | 1,822,957 |
| Molecular formula | C20H30N2O5 |
| Mr | 378.46 |
| Crystal system | Triclinic |
| Space group | P-1 |
| a, b, c (Å) | 8.0942 (4), 11.0731 (7), 11.9535 (8) |
| α, β, γ (°) | 74.538 (2), 71.071 (2), 83.527 (2) |
| V (Å3) | 976.35 (10) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| T (K) | 163 |
| Crystal size (mm) | 0.3 × 0.2 × 0.1 |
| Tmin, Tmax | 0.619, 0.745 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7404, 3725, 2578 |
| Rint | 0.030 |
| (sin θ/λ)max (Å−1) | 0.611 |
| R[F2 > 2σ(F2)], wR(F2), S | 0.04, 0.101, 0.97 |
| No. of reflections | 3725 |
| No. of parameters | 272 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Compound | Frequency (cm−1) | |||||
|---|---|---|---|---|---|---|
| vOH | Δ(vOH) | vC=O | Δ(vC=O) | vNH | Δ(NH) | |
| LID | - | - | 1662 | - | 3246 | - |
| PLG | 3217 | - | - | - | - | - |
| LID-PLG(CH2Cl2) | 3190, 3423, 3507 | −27, 206, 290 | 1623 | −39 | 3244 | −2 * |
| LID-PLG(H2O) | 3189, 3424, 3507 | −28, 207, 290 | 1621 | −41 | 3246 | 0 |
| LID-PLG(cryst) | 3189, 3424, 3507 | −28, 207, 290 | 1621 | −41 | 3244 | −2 * |
| D–H∙∙∙A | D–H | H∙∙∙A | D∙∙∙A | D–H∙∙∙A |
|---|---|---|---|---|
| N17–H17∙∙∙20 | 0.87(2) | 2.15(2) | 2.6517(2) | 116(2) |
| O1–H1∙∙∙O18 | 0.89(2) | 1.92(2) | 2.7974(2) | 171(2) |
| O3–H3∙∙∙O28 | 0.86(3) | 1.84(3) | 2.6829(2) | 171(2) |
| O5–H5∙∙∙∙O18i | 0.90(2) | 1.89(2) | 2.7578(2) | 163(2) |
| O28–H28B∙∙∙O3ii | 0.90(2) | 2.05(2) | 2.8888(2) | 155(2) |
| O28–H28A∙∙∙O5iii | 0.88(2) | 1.97(2) | 2.8288(2) | 163(2) |
| C6–H6∙∙∙O1iv | 0.95 | 2.58 | 3.5144 | 167.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magaña-Vergara, N.E.; De la Cruz-Cruz, P.; Peraza-Campos, A.L.; Martínez-Martínez, F.J.; González-González, J.S. Mechanochemical Synthesis and Crystal Structure of the Lidocaine-Phloroglucinol Hydrate 1:1:1 Complex. Crystals 2018, 8, 130. https://doi.org/10.3390/cryst8030130
Magaña-Vergara NE, De la Cruz-Cruz P, Peraza-Campos AL, Martínez-Martínez FJ, González-González JS. Mechanochemical Synthesis and Crystal Structure of the Lidocaine-Phloroglucinol Hydrate 1:1:1 Complex. Crystals. 2018; 8(3):130. https://doi.org/10.3390/cryst8030130
Chicago/Turabian StyleMagaña-Vergara, Nancy Evelyn, Porfirio De la Cruz-Cruz, Ana Lilia Peraza-Campos, Francisco Javier Martínez-Martínez, and Juan Saulo González-González. 2018. "Mechanochemical Synthesis and Crystal Structure of the Lidocaine-Phloroglucinol Hydrate 1:1:1 Complex" Crystals 8, no. 3: 130. https://doi.org/10.3390/cryst8030130
APA StyleMagaña-Vergara, N. E., De la Cruz-Cruz, P., Peraza-Campos, A. L., Martínez-Martínez, F. J., & González-González, J. S. (2018). Mechanochemical Synthesis and Crystal Structure of the Lidocaine-Phloroglucinol Hydrate 1:1:1 Complex. Crystals, 8(3), 130. https://doi.org/10.3390/cryst8030130