Synthesis, X-ray Crystal Structure and Antimicrobial Activity of Unexpected Trinuclear Cu(II) Complex from s-Triazine-Based Di-Compartmental Ligand via Self-Assembly
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Physical Measurements
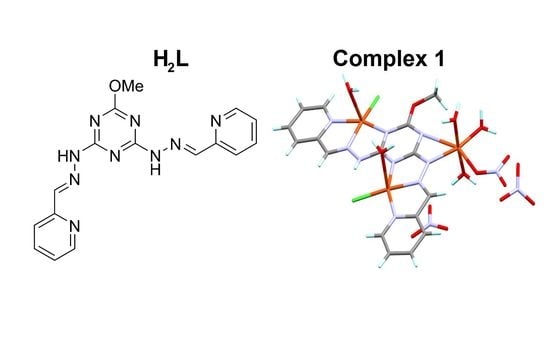
2.2. Synthesis of [Cu3(HL)(Cl)2(NO3)(H2O)5](NO3)2 (1)
2.3. X-ray Structure Determination
2.4. Hirshfeld Surface Analysis
2.5. Computational Details
2.6. Antimicrobial Experiments
2.6.1. Test Microorganism
2.6.2. Well Diffusion Method for Showing Antimicrobial Activity
2.6.3. Minimum Inhibitory Concentration (MIC) Determination
3. Results and Discussion
3.1. X-ray Structure Description
3.2. Hirshfeld Analysis
3.3. DFT Studies
3.3.1. AIM Topology Analysis
3.3.2. Natural Population Analysis
3.4. Antimicrobial Activity of H2L and its Cu(II) Complex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lehn, J.M. Programmed Chemical Systems: Multiple Subprograms and Multiple Processing/Expression of Molecular Information. Chem. Eur. J. 2000, 6, 2097–2102. [Google Scholar] [CrossRef]
- Lehn, J.M. Toward complex matter: Supramolecular chemistry and self-organization. Proc. Natl. Acad. Sci. USA 2002, 99, 4763–4768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, P.N.W.; Khoury, R.G.; Lehn, J.M.; Baum, G.; Fenske, D. Adaptive self-assembly: Environment-induced formation and reversible switching of polynuclear metallocyclophane. Chem. Eur. J. 2000, 6, 4140–4148. [Google Scholar] [CrossRef]
- Baum, G.; Constable, E.C.; Fenske, D.; Housecroft, C.E.; Kulke, T. Solvent control in the formation of mononuclear and dinuclear double-helical silver(I)-2,2′:6′,2″-terpyridine complexes. Chem. Commun. 1998, 2659–2660. [Google Scholar] [CrossRef]
- Mamula, O.; Lama, M.; Stoeckli-Evans, H.; Shova, S. Switchable Chiral Architectures Containing PrIII Ions: An Example of Solvent-Induced Adaptive Behavior. Angew. Chem. Int. Ed. 2006, 45, 4940–4944. [Google Scholar] [CrossRef] [PubMed]
- Funeriu, D.P.; Lehn, J.M.; Fromm, K.M.; Fenske, D. Multiple Expression of Molecular Information: Enforced Generation of Different Supramolecular Inorganic Architectures by Processing of the Same Ligand Information through Specific Coordination Algorithms. Chem. Eur. J. 2000, 6, 2103–2111. [Google Scholar] [CrossRef]
- Funeriu, D.P.; Rissanen, K.; Lehn, J.M. Dominant/recessive behavior in the expression of molecular information: Self-assembly of inorganic macrocyclic architectures containing coordinatively unsaturated ligands. Proc. Natl. Acad. Sci. USA 2001, 98, 10546–10551. [Google Scholar] [CrossRef] [Green Version]
- Barboiu, M.; Lehn, J.M. Dynamic chemical devices: Modulation of contraction/extension molecular motion by coupled-ion binding/pH change-induced structural switching. Proc. Natl. Acad. Sci. USA 2002, 99, 5201–5206. [Google Scholar] [CrossRef] [Green Version]
- Stadler, A.M.; Kyritsakas, N.; Lehn, J.M. Reversible folding/unfolding of linear molecular strands into helical channel-like complexes upon proton-modulated binding and release of metal ions. Chem. Commun. 2004, 2024–2025. [Google Scholar] [CrossRef]
- Ramírez, J.; Stadler, A.M.; Kyritsakas, N.; Lehn, J.M. Solvent-modulated reversible conversion of a [2×2]-grid into a pincer-like complex. Chem. Commun. 2007, 237–239. [Google Scholar] [CrossRef]
- Ramírez, J.; Stadler, A.M.; Brelot, L.; Lehn, J.M. Coordinative, conformational and motional behaviour of triazine-based ligand strands on binding of Pb(II) cations. Tetrahedron 2008, 64, 8402–8410. [Google Scholar] [CrossRef]
- Ramírez, J.; Stadler, A.M.; Harrowfield, J.M.; Brelot, L.; Huuskonen, J.; Rissanen, K.; Allouche, L.; Lehn, J.M. Coordination Architectures of Large Heavy Metal Cations (Hg2+ and Pb2+) with Bis-tridentate Ligands: Solution and Solid-State Studies. Z. Anorg. Allg. Chem. 2007, 633, 2435–2444. [Google Scholar] [CrossRef]
- Soliman, S.M.; El-Faham, A.; Elsilk, S.E.; Farooq, M. Two heptacoordinated manganese(II) complexes of giant pentadentate s-triazine bis-Schiff base ligand: Synthesis, crystal structure, biological and DFT studies. Inorg. Chim. Acta 2018, 479, 275–285. [Google Scholar] [CrossRef]
- Soliman, S.M.; El-Faham, A. Synthesis and structure diversity of high coordination number Cd(II) complexes of large s-triazine bis-Schiff base pincer chelate. Inorg. Chim. Acta 2019, 488, 131–140. [Google Scholar] [CrossRef]
- Soliman, S.M.; El-Faham, A. Synthesis and structural DFT studies of Ni(II) and Co(II) complexes with s-triazine-based di-compartmental ligand. Polyhedron 2019, 165, 162–170. [Google Scholar] [CrossRef]
- Rikagu Oxford Diffraction. CrysAlisPro; Agilent Technologies Inc.: Yarnton, Oxfordshire, UK, 2013. [Google Scholar]
- Sheldrick, G.M. Crystal structure determination with SHELXL. Acta Cryst. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer 17. University of Western Australia: Crawley, Australia, 2017. Available online: http://hirshfeldsurface.net (accessed on 1 September 2019).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. GAUSSIAN 09. Revision A02; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Dennington, R., II; Keith, T.; Millam, J. GaussView, Version 4.1; Semichem Inc.: Shawnee Mission, KS, USA, 2007. [Google Scholar]
- Adamo, C.; Barone, V. Exchange functionals with improved long-range behavior and adiabatic connection methods without adjustable parameters: The mPW and mPW1PW models. J. Chem. Phys. 1998, 108, 664–675. [Google Scholar] [CrossRef]
- Glendening, E.D.; Reed, A.E.; Carpenter, J.E.; Weinhold, F. NBO Version 3.1, CI; University of Wisconsin: Madison, WI, USA, 1998. [Google Scholar]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Barry, A.L.; Thornsberry, C. Susceptibility testing. In Manual of Clinical Microbiology; Lennette, E.H., Balows, A., Hausler, W.J., Truant, J.P., Eds.; American Society for Microbiology: Washington, DC, USA, 1980; pp. 561–574. [Google Scholar]
- Singh, V.P.; Gupta, P. Synthesis and physico-chemical studies of metal(II) complexes with diacetyl benzaldehyde acyldihydrazones and their bio-activity. J. Coord. Chem. 2008, 61, 3922–3933. [Google Scholar] [CrossRef]
- Chohan, Z.H. Ni(II), Cu(II) and Zn(II) metal chelates with some thiazole derived Schiff-bases: Their synthesis, characterization and bactericidal properties. Met. Based Drugs 1999, 6, 75–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azam, F.; Singh, S.; Khokhra, S.L.; Prakash, O. Synthesis of Schiff bases of naphtha[1,2-d]thiazol-2-amine and metal complexes of 2-(2’-hydroxy)benzylideneaminonaphthothiazole as potential antimicrobial agents. J. Zhejiang Univ. Sci. B 2007, 8, 446–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Meng, X.G.; Xiang, J.F.; Duan, Y.; Cheng, G.Z.; Ji, Z.P. Synthesis, characterization and bioactivity of four novel trinuclear copper(II) and nickel(II) complexes with pentadentate ligands derived from N-acylsalicylhydrazide. Inorg. Chim. Acta 2008, 361, 2667–2676. [Google Scholar] [CrossRef]
- Bader, R.F.W. Atoms in Molecules: A Quantum Theory; Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Matta, C.F.; Hernandez-Trujillo, J.; Tang, T.H.; Bader, R.F.W. Hydrogen-hydrogen bonding: A stabilizing interaction in molecules and crystals. Chem. Eur. J. 2003, 9, 1940–1951. [Google Scholar] [CrossRef]
- Grabowski, S.J.; Pfitzner, A.; Zabel, M.; Dubis, A.T.; Palusiak, M. Intramolecular H–H interactions for the Crystal Structures of [4-((E)-But-1-enyl)-2,6- dimethoxyphenyl]pyridine-3-carboxylate and [4-((E)-Pent-1-enyl)-2,6- dimethoxyphenyl]pyridine-3-carboxylate; DFT calculations on modeled styrene derivatives. J. Phys. Chem. B 2004, 108, 1831–1837. [Google Scholar] [CrossRef]
- Matta, C.F.; Castillo, N.; Boyd, R.J. Characterization of a closed-shell fluorine-fluorine bonding interaction in aromatic compounds on the basis of the electron density. J. Phys. Chem. A 2005, 109, 3669–3681. [Google Scholar] [CrossRef]
- Pendás, A.M.; Francisco, E.; Blanco, M.A.; Gatti, C. Bond paths as privileged exchange channels. Chem. Eur. J. 2007, 13, 9362–9371. [Google Scholar] [CrossRef]
- Bobrov, M.F.; Popova, G.V.; Tsirelson, V.G. A topological analysis of electron density and chemical bonding in cyclophosphazenes PnNnX2n (X = H, F, Cl; n = 2, 3, 4). Russ. J. Phys. Chem. 2006, 80, 584–590. [Google Scholar] [CrossRef]
- Gatti, C. Chemical bonding in crystals: New directions. Z. Kristallogr. 2005, 220, 399–457. [Google Scholar] [CrossRef]
- Gibbs, G.V.; Downs, R.T.; Cox, D.F.; Ross, N.L.; Boisen, M.B., Jr.; Rosso, K.M. Shared and closed-shell O-O interactions in silicates. J. Phys. Chem. A 2008, 112, 3693–3699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinosa, E.; Molins, E.; Lecomte, C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998, 285, 170–173. [Google Scholar] [CrossRef]










| 1 | |
|---|---|
| empirical formula | C16H24Cl2Cu3N12O15 |
| Fw | 885.99 |
| temp (K) | 120(2) K |
| λ (Å) | 0.71073 |
| crystal system | Triclinic |
| space group | P-1 |
| a (Å) | 7.1252(3) |
| b (Å) | 13.2067(5) |
| c (Å) | 16.0792(7) |
| a (deg) | 94.059(4) |
| β (deg) | 101.947(4) |
| γ (deg) | 92.712(3) |
| V (Å3) | 1473.59(11) |
| Z | 2 |
| ρcalc (Mg/m3) | 1.997 |
| μ(Mo Kα) (mm−1) | 2.418 |
| number of reflections | 14,147 |
| unique reflections | 7983 |
| GOOF (F2) * | 1.050 |
| Rint | 0.0267 |
| R1 a (I ≥ 2σ) | 0.0476 |
| wR2 b (I ≥ 2σ) | 0.1099 |
| CCDC ** | 1,965,390 |
| Bond Distances | |||
| Cu1–N2 | 1.964(3) | Cu2–Cl2 | 2.216(10) |
| Cu1–N1 | 2.041(3) | Cu2–O3 | 2.391(3) |
| Cu1–N6 | 2.062(3) | Cu3–O5 | 1.946(3) |
| Cu1–Cl1 | 2.233(9) | Cu3–O4 | 1.950(3) |
| Cu1–O1 | 2.307(3) | Cu3–O6 | 1.967(3) |
| Cu2–N8 | 1.965(3) | Cu3–N7 | 2.002(3) |
| Cu2–N9 | 2.006(3) | Cu3–N5 | 2.730(3) |
| Cu2–N4 | 2.027(3) | Cu3–O7 | 2.301(3) |
| Bond Angles | |||
| N2–Cu1–N1 | 79.11(12) | N4-Cu2–Cl2 | 103.36(9) |
| N2–Cu1–N6 | 78.19(12) | N8–Cu2–O3 | 86.79(11) |
| N1–Cu1–N6 | 157.28(12) | N9–Cu2–O3 | 99.63(11) |
| N2–Cu1–Cl1 | 168.21(9) | N4-Cu2–O3 | 87.40(11) |
| N1–Cu1–Cl1 | 96.97(9) | Cl2-Cu2–O3 | 97.89(7) |
| N6-Cu1–Cl1 | 105.13(8) | O5–Cu3–O4 | 174.67(13) |
| N2–Cu1–O1 | 86.64(11) | O5–Cu3–O6 | 84.02(19) |
| N1–Cu1–O1 | 87.67(11) | O4–Cu3–O6 | 90.67(19) |
| N6-Cu1–O1 | 91.86(11) | O5–Cu3–N7 | 90.58(12) |
| Cl1-Cu1–O1 | 104.40(7) | O4–Cu3–N7 | 94.18(12) |
| N8–Cu2–N9 | 80.46(12) | O6-Cu3–N7 | 157.47(18) |
| N8–Cu2–N4 | 78.43(11) | O5–Cu3–O7 | 90.82(13) |
| N9–Cu2–N4 | 157.32(12) | O4–Cu3–O7 | 90.81(14) |
| N8–Cu2–Cl2 | 175.03(9) | O6-Cu3–O7 | 103.30(17) |
| N9–Cu2–Cl2 | 97.04(9) | N7-Cu3–O7 | 98.62(11) |
| D–H–A | d(D–H) | d(H–A) | d(D–A) | <(DHA) |
|---|---|---|---|---|
| O1–H1B–O14#1 | 0.84 | 2.15 | 2.929(4) | 153.1 |
| O1–H1B–O15#1 | 0.84 | 2.43 | 3.184(4) | 149.1 |
| O3–H3A–O8#2 | 0.87 | 2.06 | 2.910(4) | 165.2 |
| O3–H3B–O15#1 | 0.93 | 2.05 | 2.895(5) | 150.0 |
| O4–H4A–O8#2 | 0.86 | 1.91 | 2.728(4) | 159.9 |
| O4–H4B–O10#1 | 0.89 | 1.92 | 2.766(4) | 157.2 |
| O5–H5A–O10 | 0.83 | 1.82 | 2.641(4) | 172.0 |
| O5–H5B–O14 | 0.87 | 1.85 | 2.715(5) | 174.2 |
| O6-H6B–O11#1 | 0.89 | 1.87 | 2.698(6) | 153.4 |
| C4–H4–O9#3 | 0.95 | 2.3 | 3.192(5) | 156.6 |
| C6–H6–O3#4 | 0.95 | 2.26 | 3.199(4) | 168.1 |
| Atom or Ligand Group | Charge |
|---|---|
| Cu1 | 0.8297 |
| H2O(1) | 0.0805 |
| Cl1 | −0.5191 |
| Cu2 | 0.8378 |
| H2O(3) | 0.0682 |
| Cl2 | −0.5905 |
| Cu3 | 1.0141 |
| H2O(4) | 0.1602 |
| H2O(5) | 0.1892 |
| H2O(6) | 0.1717 |
| N(10)O3− | −0.8550 |
| N(11)O3− | −0.9484 |
| N(12)O3− | −0.9243 |
| HL− | 0.4860 |
| Target microbes | Cu(II) Complex | H2L | Gentamicin a |
|---|---|---|---|
| S. aureus | 16 | 34 | 34 |
| Streptococcus epidermidis | 16 | 22 | 32 |
| Enterococcus faecalis | 9 | 13 | 21 |
| E. coli | 10 | 14 | 21 |
| S. typhi | 12 | 11 | 22 |
| Pseudomonas aeruginosa | 17 | 17 | 19 |
| C. albicans | 7 | 7 | - |
| Microbes | Complex 1 a | H2L a |
|---|---|---|
| E. coli | 0.375(0.563) | 0.750 (1.125) |
| S. epidermidis | 0.188 (0.282) | 0.375 (0.563) |
| C. albicans | 0.188 (0.282) | 0.750 (1.125) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliman, S.M.; Lasri, J.; Haukka, M.; Sholkamy, E.N.; Al-Rasheed, H.H.; El-Faham, A. Synthesis, X-ray Crystal Structure and Antimicrobial Activity of Unexpected Trinuclear Cu(II) Complex from s-Triazine-Based Di-Compartmental Ligand via Self-Assembly. Crystals 2019, 9, 661. https://doi.org/10.3390/cryst9120661
Soliman SM, Lasri J, Haukka M, Sholkamy EN, Al-Rasheed HH, El-Faham A. Synthesis, X-ray Crystal Structure and Antimicrobial Activity of Unexpected Trinuclear Cu(II) Complex from s-Triazine-Based Di-Compartmental Ligand via Self-Assembly. Crystals. 2019; 9(12):661. https://doi.org/10.3390/cryst9120661
Chicago/Turabian StyleSoliman, Saied M., Jamal Lasri, Matti Haukka, Essam N. Sholkamy, Hessa H. Al-Rasheed, and Ayman El-Faham. 2019. "Synthesis, X-ray Crystal Structure and Antimicrobial Activity of Unexpected Trinuclear Cu(II) Complex from s-Triazine-Based Di-Compartmental Ligand via Self-Assembly" Crystals 9, no. 12: 661. https://doi.org/10.3390/cryst9120661
APA StyleSoliman, S. M., Lasri, J., Haukka, M., Sholkamy, E. N., Al-Rasheed, H. H., & El-Faham, A. (2019). Synthesis, X-ray Crystal Structure and Antimicrobial Activity of Unexpected Trinuclear Cu(II) Complex from s-Triazine-Based Di-Compartmental Ligand via Self-Assembly. Crystals, 9(12), 661. https://doi.org/10.3390/cryst9120661