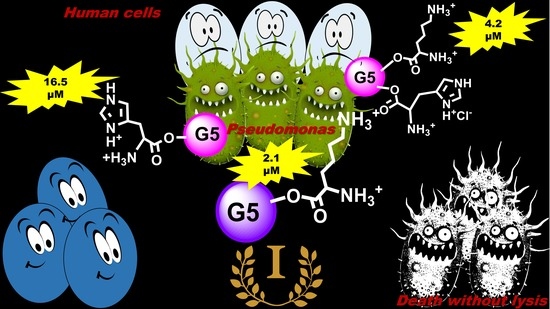
Antibacterial Activity of Non-Cytotoxic, Amino Acid-Modified Polycationic Dendrimers against Pseudomonas aeruginosa and Other Non-Fermenting Gram-Negative Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Antimicrobial Assays
2.3. Killing Curves
2.4. Evaluation of the Antimicrobial Effect of G5K by Turbidimetric Studies
3. Results and Discussion
3.1. Selection of Positively Charged Dendrimers G5K, G5H, and G5HK
3.2. Antimicrobial Activities of G5K, G5H, and G5HK
3.3. Time-Killing Curves
3.4. Effect of G5K on the Growth Curve of P. aeruginosa, A. baumannii, and S. maltophilia
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug Resistant Bacterial Infections, Including Tuberculosis; WHO: Geneva, Switzerland, 2017; Available online: https://www.who.int/medicines/areas/rational_use/PPLreport_2017_09_19.pdf?ua=1 (accessed on 12 August 2020).
- World Health Organization (WHO). No Time to Wait: Securing the Future from Drug-Resistant Infections. Report to the Secretary-General of the United Nations; Interagency Coordination Group on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2019; Available online: https://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_final_report_EN.pdf?ua=1 (accessed on 12 August 2020).
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.D.; Won, H.S.; Kim, J.H.; Mishig-Ochir, T.; Lee, B.J. Antimicrobial peptides for therapeutic applications: A review. Molecules 2012, 17, 12276–12286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, Y.J.; Romanowski, E.G.; McDermott, A.M. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. 2005, 30, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Landman, D.; Georgescu, C.; Martin, D.A.; Quale, J. Polymyxins revisited. Clin. Microbiol. Rev. 2008, 21, 449–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karaiskos, I.; Souli, M.; Galani, I.; Giamarellou, H. Colistin: Still a lifesaver for the 21st century? Expert Opin. Drug Metab. Toxicol. 2017, 13, 59–71. [Google Scholar] [CrossRef]
- Alfei, S.; Schito, A. Positively charged polymers as promising devices against multidrug resistant Gram-Negative bacteria: A review. Polymers 2020, 12, e1195. [Google Scholar] [CrossRef]
- Kang, H.K.; Kim, C.; Seo, C.H.; Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): A patent review. J. Microbiol. 2017, 55, 1–12. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Yang, L.; Gordon, V.D.; Trinkle, D.; Schmidt, N.W.; Davis, M.A.; Devries, C.; Som, A.; Cronan, J.E.; Tew, G.N.; Wong, G.C.L. Mechanism of a prototypical synthetic membrane-active antimicrobial: Efficient hole-punching via interaction with negative intrinsic curvature lipids. Proc. Natl. Acad. Sci. USA 2008, 105, 20595–20600. [Google Scholar] [CrossRef] [Green Version]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K.; et al. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019, 10, e4538. [Google Scholar] [CrossRef]
- Alfei, S.; Signorello, M.G.; Schito, A.; Catena, S.; Turrini, F. Reshaped as polyester-based nanoparticles, gallic acid inhibits platelet aggregation, reactive oxygen species production and multi-resistant Gram-positive bacteria with an efficiency never obtained. Nanoscale Adv. 2019, 1, 4148–4157. [Google Scholar] [CrossRef] [Green Version]
- Alfei, S.; Catena, S.; Turrini, F. Biodegradable and biocompatible spherical dendrimer nanoparticles with a gallic acid shell and a double-acting strong antioxidant activity as potential device to fight diseases from “oxidative stress”. Drug Deliv. Transl. Res. 2020, 10, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Alfei, S.; Marengo, B.; Domenicotti, C. Polyester-based dendrimer nanoparticles combined with etoposide have an improved cytotoxic and pro-oxidant effect on human neuroblastoma cells. Antioxidants 2020, 9, e50. [Google Scholar] [CrossRef] [Green Version]
- Alfei, S.; Marengo, B.; Zuccari, G.; Turrini, F.; Domenicotti, C. Dendrimer nanodevices and gallic acid as novel strategies to fight chemoresistance in neuroblastoma cells. Nanomaterials 2020, 10, e1243. [Google Scholar] [CrossRef]
- Avval, M.M.; Murthy, S.V.; Shashikanth, S. Synthesis and antimicrobial activity evaluation of poly ethylene imine (pei) dendrimer modified with 1,3,4 oxadiazole derivatives. Res. J. Pharm. Biol. Chem. Sci. 2014, 5, 678–683. [Google Scholar]
- Gibney, K.A.; Sovadinova, I.; Lopez, A.I.; Urban, M.; Ridgway, Z.; Caputo, G.A.; Kuroda, K. Poly(ethylene imine)s as antimicrobial agents with selective activity. Macromol. Biosci. 2012, 12, 1279–1289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pakrudheen, I.; Banu, A.N.; Murugan, E. Cationic amphiphilic dendrimers with tunable hydrophobicity show in vitro activity. Environ. Chem. Lett. 2018, 16, 1513–1519. [Google Scholar] [CrossRef]
- Kannan, R.; Prabakaran, P.; Basu, R.; Pindi, C.; Senapati, S.; Muthuvijayan, V.; Prasad, E. Mechanistic study on the antibacterial activity of self-assembled poly(aryl ether)-based amphiphilic dendrimers. ACS Appl. Bio Mater. 2019, 2, 3212–3224. [Google Scholar] [CrossRef] [Green Version]
- Neelgund, G.M.; Oki, A.; Luo, Z. Antimicrobial activity of CdS and Ag2S quantum dots immobilized on poly(amidoamine) grafted carbon nanotubes. Colloids Surfaces B Biointerfaces 2012, 100, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Pires, J.; Siriwardena, T.N.; Stach, M.; Tinguely, R.; Kasraian, S.; Luzzaro, F.; Leib, S.L.; Darbre, T.; Reymond, J.-L.; Endimiani, A. In Vitro activity of the novel antimicrobial peptide dendrimer G3KL against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015, 59, 7915–7918. [Google Scholar] [CrossRef] [Green Version]
- Young, A.W.; Liu, Z.; Zhou, C.; Totsingan, F.; Jiwrajka, N.; Shi, Z.; Kallenbach, N.R. Structure and antimicrobial properties of multivalent short peptides. Med. Chem. Comm. 2011, 2, 308–314. [Google Scholar] [CrossRef]
- Siriwardena, T.N.; Capecchi, A.; Gan, B.-H.; Jin, X.; He, R.; Wei, D.; Ma, L.; Köhler, T.; van Delden, C.; Javor, S.; et al. Optimizing antimicrobial peptide dendrimers in chemical Space. Angew. Chem. Int. Ed. 2018, 57, 8483–8487. [Google Scholar] [CrossRef]
- Siriwardena, T.N.; Lüscher, A.; Köhler, T.; van Delden, C.; Javor, S.; Reymond, J.-L. Antimicrobial peptide dendrimer chimera. Helvetica Chim. Acta 2019, 102. [Google Scholar] [CrossRef]
- Mintzer, M.A.; Dane, E.L.; O’Toole, G.A.; Grinstaff, M.W. Exploiting dendrimer multivalency to combat emerging and re-emerging infectious diseases. Mol. Pharm. 2012, 9, 342–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfei, S.; Castellaro, S. Synthesis and characterization of polyester-based dendrimers containing peripheral arginine or mixed amino acids as potential vectors for gene and drug delivery. Macromol. Res. 2017, 25, 1172–1186. [Google Scholar] [CrossRef]
- Alfei, S.; Catena, S. Synthesis and characterization of versatile amphiphilic dendrimers peripherally decorated with positive charged amino acids. Polym. Int. 2018, 67, 1572–1584. [Google Scholar] [CrossRef]
- Alfei, S.; Catena, S. Synthesis and characterization of fourth generation polyester-based dendrimers with cationic amino acids-modified crown as promising water soluble biomedical devices. Polym. Adv. Technol. 2018, 29, 2735–2749. [Google Scholar] [CrossRef]
- Alfei, S.; Castellaro, S.; Taptue, G.B. Synthesis and NMR characterization of dendrimers based on 2, 2-bis-(hydroxymethyl)-propanoic acid (bis-HMPA) containing peripheral amino acid residues for gene transfection. Org. Commun. 2017, 10, 144–177. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/ast_of_bacteria/ (accessed on 31 July 2020).
- Schito, A.M.; Piatti, G.; Stauder, M.; Bisio, A.; Giacomelli, E.; Romussi, G.; Pruzzo, C. Effects of demethylfruticuline A and fruticuline A from Salvia corrugata Vahl. on biofilm production in vitro by multiresistant strains of Staphylococcus aureus, Staphylococcus epidermidis and Enterococcus faecalis. Int. J. Antimicrob. Agents 2011, 37, 129–134. [Google Scholar] [CrossRef] [Green Version]
- Dalgaard, P.; Ross, T.; Kamperman, L.; Neumeyer, K.; McMeekin, T.A. Estimation of bacterial growth rates from turbidimetric and viable count data. Int. J. Food Microbiol. 1994, 23, 391–404. [Google Scholar] [CrossRef]
- Jain, A.; Duvvuri, L.S.; Farah, S.; Beyth, N.; Domb, A.J.; Khan, W. Antimicrobial polymers. Adv. Healthc. Mater. 2014, 3, 1969–1985. [Google Scholar] [CrossRef] [PubMed]
- Gholami, M.; Mohammadi, R.; Arzanlou, M.; Akbari Dourbash, F.; Kouhsari, E.; Majidi, G.; Mohseni, S.M.; Nazari, S. In vitro antibacterial activity of poly (amidoamine)-G7 dendrimer. BMC Infect. Dis. 2017, 17, 395. [Google Scholar] [CrossRef] [PubMed]
- Glukhov, E.; Stark, M.; Burrows, L.L.; Deber, C.M. Basis for selectivity of cationic antimicrobial peptides for bacterial versus mammalian membranes. J. Biol. Chem. 2005, 280, 33960–33967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganewatta, M.S.; Tang, C. Controlling macromolecular structures towards effective antimicrobial polymers. Polymer 2015, 63, A1–A29. [Google Scholar] [CrossRef]
- Stenström, P.; Hjorth, E.; Zhang, Y.; Andrén, O.C.J.; Guette-Marquet, S.; Schultzberg, M.; Malkoch, M. Synthesis and in vitro evaluation of monodisperse amino-functional polyester dendrimers with rapid degradability and antibacterial properties. Biomacromolecules 2017, 18, 4323–4330. [Google Scholar] [CrossRef]
- Stach, M.; Siriwardena, T.N.; Köhler, T.; Van Delden, C.; Darbre, T.; Reymond, J.-L. Combining topology and sequence design for the discovery of potent antimicrobial peptide dendrimers against multidrug-resistant pseudomonas aeruginosa. Angew. Chem. Int. Ed. 2014, 53, 12827–12831. [Google Scholar] [CrossRef]
- Niederhafner, P.; Bednárová, L.; Buděšínský, M.; Šafařík, M.; Ehala, S.; Ježek, J.; Borovičková, L.; Fučík, V.; Čeřovský, V.; Slaninová, J. Melectin MAPs: The influence of dendrimerization on antimicrobial and hemolytic activity. Amino Acids 2010, 39, 1553–1561. [Google Scholar] [CrossRef]
- Yang, X.; Hu, K.; Hu, G.; Shi, D.; Jiang, Y.; Hui, L.; Zhu, R.; Xie, Y.; Yang, L. Long hydrophilic-and-cationic polymers: A different pathway toward preferential activity against bacterial over mammalian membranes. Biomacromol. 2014, 15, 3267–3277. [Google Scholar] [CrossRef]
- Rivera, M.; Bryan, L.E.; Hancock, R.E.V.; Mc Groarty, E.J. Heterogeneity of lipopolysaccharides from Pseudomonas aeruginosa: Analysis of lipopolysaccharide chain length. J. Bacteriol. 1988, 170, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, I.E.; Kintz, E.N.; Porter, L.A.; Goldberg, J.B.; Burnham, N.A.; Camesano, T.A. Relating the physical properties of Pseudomonas aeruginosa lipopolysaccharides to virulence by atomic force microscopy. J. Bacteriol. 2011, 193, 1259–1266. [Google Scholar] [CrossRef] [Green Version]
- Gough, N.R. Stressing bacteria to death. Sci. Signal. 2011, 4, e164. [Google Scholar] [CrossRef]



| Strains | G5HK 1 mg/L; (mM) | G5H 1 mg/L; (mM) | G5K 1 mg/L; (mM) |
|---|---|---|---|
| S. aureus 119 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| S. aureus 118 (MRSA) | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| S. epidermidis 199 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| S. epidermidis 197 (MRSE) | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| E. faecalis 124 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| E. faecalis 120 (VRE) | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| E. faecium 127 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| E. faecium 118 (VRE) | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| E. coli 123 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| E. coli 133 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| K. pneumonia 236 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| K. pneumoniae 237 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| P. mirabilis 155 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| P. aeruginosa 208 | 256; (0.0084) | 1024; (0.0332) | 64; (0.0021) |
| P. aeruginosa 209 | 128; (0.0042) | 512; (0.0166) | 64; (0.0021) |
| P. aeruginosa 230 | 256; (0.0084) | 1024; (0.0332) | 64; (0.0021) |
| P. aeruginosa 247 | 256; (0.0084) | 1024; (0.0332) | 64; (0.0021) |
| P. aeruginosa 248 | 256; (0.0084) | 1024; (0.0332) | 64; (0.0021) |
| P. aeruginosa 249 | 128; (0.0042) | 1024; (0.0332) | 64; (0.0021) |
| P. aeruginosa 253 | 512; (0.0168) | 512; (0.0166) | 64; (0.0021) |
| P. aeruginosa 256 | 256; (0.0084) | 1024; (0.0332) | 64; (0.0021) |
| P. aeruginosa 259 | 512; (0.0168) | 1024; (0.0332) | 64; (0.0021) |
| P. aeruginosa 265 | >1024; (>0.0334) | >1024; (>0.0332) | >1024; (>0.0329) |
| P. aeruginosa ATCC 27853 | 256; (0.0084) | 1024; (0.0332) | 64; (0.0021) |
| P. putida 262 | 64; (0.0021) | 512; (0.0166) | 32; (0.0010) |
| P. fluorescens 263 | 32; (0.0010) | 256; (0.0083) | 16; (0.0005) |
| P. straminea A4 | 32; (0.0010) | 256; (0.0083) | 32; (0.0010) |
| S. maltophilia 11 | 128; (0.0042) | 1024; (0.0332) | 64; (0.0021) |
| S. maltophilia 16 | 256; (0.0084) | 512; (0.0166) | 128; (0.0042) |
| S. maltophilia 18 | 64; (0.0021) | 256; (0.0083) | 32; (0.00105) |
| S. maltophilia 19 | 256; (0.0084) | 512; (0.0166) | 128; (0.0042) |
| A. baumanni 23 | 128; (0.0042) | 512; (0.0166) | 64; (0.0021) |
| A. baumanni 24 | 128; (0.0042) | 256; (0.0083) | 32; (0.00105) |
| A. baumanni 25 | 256; (0.0084) | 512; (0.0166) | 64; (0.0021) |
| A. baumanni 27 | 128; (0.0042) | 1024; (0.0332) | 64; (0.0021) |
| A. pittii 272 | 64; (0.0021) | 256; (0.0083) | 64; (0.0021) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schito, A.M.; Alfei, S. Antibacterial Activity of Non-Cytotoxic, Amino Acid-Modified Polycationic Dendrimers against Pseudomonas aeruginosa and Other Non-Fermenting Gram-Negative Bacteria. Polymers 2020, 12, 1818. https://doi.org/10.3390/polym12081818
Schito AM, Alfei S. Antibacterial Activity of Non-Cytotoxic, Amino Acid-Modified Polycationic Dendrimers against Pseudomonas aeruginosa and Other Non-Fermenting Gram-Negative Bacteria. Polymers. 2020; 12(8):1818. https://doi.org/10.3390/polym12081818
Chicago/Turabian StyleSchito, Anna Maria, and Silvana Alfei. 2020. "Antibacterial Activity of Non-Cytotoxic, Amino Acid-Modified Polycationic Dendrimers against Pseudomonas aeruginosa and Other Non-Fermenting Gram-Negative Bacteria" Polymers 12, no. 8: 1818. https://doi.org/10.3390/polym12081818
APA StyleSchito, A. M., & Alfei, S. (2020). Antibacterial Activity of Non-Cytotoxic, Amino Acid-Modified Polycationic Dendrimers against Pseudomonas aeruginosa and Other Non-Fermenting Gram-Negative Bacteria. Polymers, 12(8), 1818. https://doi.org/10.3390/polym12081818