1. Introduction
Osteosarcoma (OS) is a highly aggressive bone malignancy that primarily affects adolescents and young adults. The standard-of-care regimen is based on local surgical control of the disease combined with high-dose chemotherapy. First-line conventional drugs include methotrexate, doxorubicin, cisplatin, and possibly ifosfamide (MAPI) or etoposide [
1,
2]. Despite the various attempts to ameliorate clinical outcomes, the survival rate has not improved over the past three decades; indeed, the 5-year survival rate for localized OS is still 60–70%, whereas survival for patients who do not respond to first line-therapy and develop metastases drops to 20–30% [
1,
3,
4,
5]. The occurrence of drug resistance [
6,
7] and development of metastasis (lung metastasis in 80% of the cases) [
8] are strongly associated with poor prognosis, and represent critical hallmarks in OS progression [
9,
10]. In particular, chemoresistant and/or metastatic OS are heterogeneous, genetically complex, and therapeutically challenging because such cases cannot be treated by surgery and are refractory to conventional chemotherapy. Many new therapeutic options, including targeted therapy, immunotherapy, and intensive chemotherapy with peripheral blood stem cell reinfusion, are under investigation, but none have yet shown a major change in overall survival [
11,
12,
13,
14].
A better understanding of the mechanisms of drug resistance and metastasis formation is, therefore, mandatory to find new therapeutic targets. In addition, the clarification of the interactions between OS cells and the complex microenvironment of bone and bone marrow is highly required, since the tumor microenvironment plays a critical role in regulating cancer invasion and drug resistance [
15]. In this respect, three-dimensional (3D) cell cultures offer the opportunity to simulate microenvironmental conditions, such as hypoxia and nutrient gradients, to co-culture two or more different cell types, and recapitulate native cell architecture and stiffness, a condition known to affect the cell behavior also in terms of gene expression profile and drug sensitivity. Moreover, 3D models allow to observe the invasive processes in real-time, interrogate the molecular pathways involved in cell migration and proliferation, and screen compounds with the potential anticancer activity in a condition that better represents in vivo tissues than conventional 2D cultures. Consequently, 3D cell cultures are increasingly becoming popular tools for basic research and drug screening. However, they do often rely on manual, multi-step fabrication techniques, which may limit the acquisition of repeatable and standardized results. In this respect, advanced bio-fabrication techniques, such as bioprinting, represent promising innovative approaches for developing physiologically relevant tissues or disease models. Through the simultaneous printing of cells, biomaterials, and bioactive cues with accurate spatial control, bioprinting allows the production of highly complex 3D microtissues, and enables the creation of models through an automated, repeatable approach, with the virtue of providing accurate control for in vitro biological and/or drug screening assays [
16,
17]. In the context of OS, 3D printing can produce novel bone tissue engineering scaffolds with customized shape, architecture, favorable macro-microstructure, mechanical strength, and cellular components [
18].
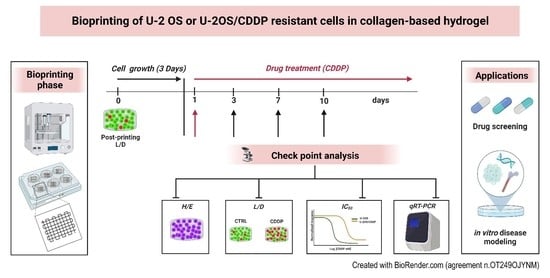
In this study, we present an innovative preclinical 3D OS model through a 3D bioprinting platform using a natural derived, collagen-based bioink. Hydrogels have been widely used as scaffolds for tissue engineering because of their excellent biocompatibility. Among hydrogel materials (such as alginate, fibrinogen, gelatin, hyaluronic acid, and collagen), collagen was here favored to better mimic the natural bone mechanical environment. We introduced the 3D bioprinting process and the method of construction of the 3D tumor models, verified whether these scaffolds were biocompatible in maintaining cell viability, and tested their biological potential for evaluating cancer cell proliferation and spreading compared to 2D conditions. The results of biological characterization of cell proliferation, invasion, and chemosensitivity together with cell expression of matrix metalloproteinases (MMPs) and biomarkers of resistance to cisplatin (CDDP) for the printed 3D OS tumor models are shown.
2. Materials and Methods
2.1. OS Cell Lines
We used U-2 OS and its CDDP-resistant variant, U-2OS/CDDP 1 μg, to develop a simplified 3D OS model. The U-2OS/CDDP 1 μg cell line was established by exposing the CDDP-sensitive U-2 OS purchased from the American Type Culture Collection (ATCC) to step-by-step increases in CDDP (#P4394, Sigma Aldrich St Louis, MO, USA; concentrations as previously described [
19]). Cell lines were tested for mycoplasma contamination every 3 months (#LT07-318, MycoAlert Mycoplasma Detection Kit, Lonza, Basel, Switzerland) and were authenticated (STR profiling) by analysis of the following loci: AMEL, D3S1358, TH01, D21S11, D18S51, D10S1248, D1S1656, D2S1338, D16S539, D22S1045, VWA, D8S1179, FGA, D2S441, D12S391, D19S433, and SE33 (last control July 2020; POWERPLEX ESX 17 Fast System, Promega, Madison, WI, USA). All cell lines were immediately amplified to constitute liquid nitrogen stocks and were never passaged for more than 1 month upon thawing. Cells were cultured with Iscove’s Modified Dulbecco’s Medium (#ECB2072L, IMDM, Euroclone, Milan, Italy) containing 10% heat-inactivated fetal bovine serum (FBS, #ECS0180L, Euroclone Milan, Italy), penicillin (100 units/mL), and streptomycin (100 μg/mL) (#ECB3001D, Euroclone Milan, Italy) maintained at 37 °C in a humidified 5% CO
2 atmosphere.
2.2. Design and Fabrication of a 3D Osteosarcoma Model
The development of the 3D in-vitro model foresaw the design and fabrication of cell-laden 3D scaffolds using a bioprinting platform. For the first phase, a dedicated CAD software (BioCAD, RegenHU, CH) was used to design the 3D structures, characterized by a 10 × 10 mm square basis and a total height of 2 mm. A 0/90° infill pattern was selected as the internal microarchitecture, with a fiber diameter of 300 µm and pore size of 1700 µm. To guarantee an optimal adhesion within the different deposited layers, a layer height equal to 200 µm, approximately 70% of the fiber diameter, was selected, leading to the stacking of 10 layers to achieve the desired construct height. The design software enabled us to set the printing process to be automatically replicated within 6-well plates for higher throughput fabrication. The first phase of the fabrication of the 3D OS model included the preparation of the cell-laden bioink, according to the protocol provided by the manufacturer.
First, 1 mL of high-density collagen bioink (80 mg/mL Viscoll, Imtek, RU) was neutralized with a solution composed of 100 µL buffer (Imtek, RU) and 400 µL FBS. Preliminary analyses were carried out to test the biocompatibility of the buffer on OS cell lines through Trypan Blue solution (#T8154-100ML, Sigma-Aldrich St Louis, MO, USA) cell count at 1, 3, 6, and 24 h. U-2 OS and U-2OS/CDDP 1 μg cells were trypsinized, counted, and re-suspended at 2 × 106 cells/mL in 300 µL of FBS. The suspension was then mixed with the neutralized collagen ink through a Luer-lock coupler (Sarstedt AG & Co. KG, Numbrecht, DE), performing 20 mixing iterations by gently and alternately pushing the two syringe plungers, as per the protocols provided by the manufacturer. Finally, the cell-laden bioink was loaded into the bioprinter cartridge.
The 3D bioprinting process was performed through a 3D Discovery platform (RegenHU, CH). High-precision microvalve-based dispensing technology, used in contact mode with a dedicated 300 µm needle, was chosen to maximize the printing fidelity of the low viscosity bioink. The printhead was kept at room temperature (RT) throughout the experiment, whereas the build zone temperature was set at 37 °C to favor physical gelation of the collagen bioink and improve the shape retention, as well as the stacking behavior, of the deposited fibers. The process was automatedly replicated in sterile ultra-low attachment 6 well-plates (#3471, Corning Inc., New York, NY, USA). Sterility was guaranteed by the printing platform being integrated in a class II laminar flow hood. As technical control, we used cell lines mixed with the collagen bioink, which did not undergo the printing process, to evaluate the effect of the procedure and the selected parameters on cell viability. Cell-laden hydrogels were kept in culture in the same experimental conditions reported above for up to 10 days by changing the culture media twice per week (
Video S1). During the culture time, cells in hydrogels were observed through a phase-contrast microscope before all the scheduled assessments.
2.3. Analysis of Biological Parameters in 3D Conditions
2.3.1. Cell Viability and Morphology
The viability of OS cells in the cell-laden bioinks was determined by using a Live/Dead® cell viability kit (#L3224, Invitrogen, Waltham, MA, USA) following the manufacturer’s instructions. Briefly, constructs were rinsed with phosphate-buffered saline (PBS) (Sigma-Aldrich, St. Louis, MO, USA) and then incubated with calcein-AM and ethidium bromide solution (#L3224, Invitrogen, Waltham, MA, USA) for 30′ at 37 °C. After three washing steps with PBS, constructs were visualized with the epi-fluorescent microscope, Eclipse 90i (Nikon, Melville, NY, USA), using the fluorescein isothiocyanate (FITC) and tetramethyl rhodamine (TRITC) filters to evaluate the viable (green) and dead (red) cells, respectively. Beyond 2D microscopic acquisition, we performed Z-stack imaging in real-time from the top to the bottom for 3D reconstruction of some areas of constructs through Eclipse 90i software (Nikon, Melville, NY, USA). In parallel, viability and metabolic activities of OS cells in the 3D bioinks were assessed by Alamar BlueTM Cell Viability Reagent (#DAL1100, Invitrogen, Thermo Scientific, Waltham, MA, USA) at day 3, 7, and 10. The 3D constructs were incubated with complete medium supplemented with 10% Alamar BlueTM (Invitrogen, Thermo Scientific, Waltham, MA, USA) for 4 h a 37 °C in humidified 5% CO2. After the incubation, 100 µL of the solution was transferred to a 96-well plate, and the fluorescence signals were measured at an excitation length of 530–560 nm and an emission length of 590 nm by a Glomax Multi Detection System (Promega Italia S.r.l, Milano, Italy). A morphological evaluation of OS cells grown in the 3D bio-printed constructs was also performed at day 3, 7, and 10 after cell seeding. Three-dimensional constructs were embedded in the Tissue-Tek® optimal cutting temperature (OCT) compound (#4583, Sakura Finetech, CA, USA), and then snap-frozen in liquid nitrogen. After cutting with the cryostat 12-μm thick sections, slides were fixed with paraformaldehyde solution 4% (#P6148, Sigma Aldrich St Louis, MO, USA), and then processed for Gill III Hematoxylin-Eosin (H/E) (#05-M06015, Bio-Optica, Milano, Italy) staining to evaluate the morphology of U-2 OS and U-2OS/CDDP 1 μg and cancer cell crosstalk with the collagen-based hydrogen at different time points.
2.3.2. Cell Migration and Proliferation
Morphological evaluation of the invasive capabilities of OS cells in the bioprinted collagen scaffolds was performed after H/E staining of sections from 3D constructs embedded in OCT over 3–10 days. Evaluation of the expression of MMPs and the biomarker of chemoresistance was performed on day 10. The expression of Ki-67 was also determined as a measure of cell proliferation.
2.3.3. Drug Response
Two 106 U-2 OS or U-2OS/CDDP 1-ug cells were bio printed, cultured in hydrogels for 3 days, and checked for morphology and vitality before being exposed to different doses of CDDP (U-2 OS: 300 ng/mL, 1 μg/mL, and 3 μg/mL; U-2OS/CDDP 1 μg: 1 μg/mL, 3 μg/mL, and 10 μg/mL). Drug efficacy was evaluated over 3–10 days using either Live and Dead or Alamar Blue assays. IC50 (concentration required to inhibit cell proliferation by 50%) values were calculated from the linear transformation of dose–response curves.
2.4. Analysis of Biological Parameters in 2D Conditions
Cells were plated into 96-well plates (2000 cells/well) in standard medium and cultured for 24 h, before they were treated with CDDP for an additional 96 h, roughly equivalent to at least two doubling times of each cell line. Drug efficacy was assessed with the TACS® MTT Cell Proliferation Assay kit (#4890-25-K, Trevigen, Inc., Gaithersburg, MD, USA) according to the manufacturer’s instructions.
For evaluating the expression of Ki-67, MMPs, and/or biomarkers of chemoresistance, 2 × 105 cells were plated in 6-well dishes. At day 3 from cell seeding, when cells are in active growth, Ki-67 expression was assessed by immunofluorescence, and total RNA was extracted for gene evaluation by quantitative PCR (q-PCR).
2.5. Immunofluorescence
For 3D conditions, OS cell-laden bioinks were kept in culture for 3–10 days, and each sample was embedded in the OCT compound and snap-frozen. Slides were fixed in paraformaldehyde solution 4% (Kaltek srl, Padova, IT), and hydrated with Tris-buffered saline (TBS, #T5912, Sigma-Aldrich, St Louis, MO, USA) and the albumin solution 1% (#A4503, Sigma-Aldrich St Louis, MO, US) to block the non-specific antigenic sites for 30′ at RT. Then, samples were incubated with the mouse monoclonal anti-human antibodies, Ki-67 (clone UMAB107, #UM800033; Origene, Rockville, MD, USA; 10 µg/mL) or MMP-13 (Clone 87512, #MAB511, R&D Systems, CA, USA; 5 μg/mL). Following the washing steps in TBS, sections were incubated with the goat anti-mouse secondary antibody conjugated with FITC (#31569; Thermo Fisher Scientific, Waltham, MA, USA; 15 µg/mL) for 1 h at 4 °C. Negative controls were carried out either by avoiding the use of the primary antibodies or with isotype control. After three washing steps, samples were mounted with the anti-fading solution containing 4′,6-diamidine-2-phenylindole (DAPI, #D9542, Sigma St Louis, MO, USA) for nuclear staining. Section assessment was performed through the epi-fluorescent microscope, Eclipse 90i (Nikon). Ten microscopic fields (40× magnification) were used to perform a semi-quantitative analysis of immunofluorescence for Ki-67 and MMP-13 markers. Technical triplicates were used for each time point.
For 2D conditions, cells were fixed with 4% paraformaldehyde solution, and then immunoassayed as for 3D sections.
2.6. RNA Extraction and Quantitative PCR
The total RNA from OS cells constructs was extracted with TRIzol Reagent (#15596018; Thermo Fisher Scientific—Life Technologies, Grand Island, NY, USA) following the manufacturer’s instructions. RNA quality and quantity were assessed by NanoDrop analysis (NanoDrop ND1000, Thermo Scientific, MA, USA). The total RNA from each sample was reverse-transcribed into complementary DNA (cDNA) using a High-Capacity cDNA Reverse Transcription Kit (#4368813; Thermo Fisher Scientific—Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s protocols. Quantitative reverse transcriptase-polymerase chain reaction (qPCR) was performed on a ViiATM 7 System (Applied Biosystems, Waltham, MA, USA) using TaqMan Universal PCR Master Mix (#4304437, Thermo Fisher Scientific-Applied Biosystems) and SYBR Green PCR Master Mix (#4312704, Thermo Fisher Scientific—Applied Biosystem, Waltham, MA, USA). Predesigned TaqMan assays (#4351370, Thermo Fisher Scientific—Applied Biosystems, Waltham, MA, USA) were used for the following genes: GAPDH (Hs99999905_m1), GSTP1 (Hs00168310_m1), XPA (Hs00166045_m1), ERCC1 (Hs01012158_m1), ERCC2 (Hs00361161_m1), ERCC5 (Hs01557031_m1), ERCC4 (Hs00193342_m1), MMP-1 (Hs00899658_m1), MMP-9 (Hs00234579_m1). We used the following primers for MMP-13 (forward, 5′-TCA CGA TGG CAT TGC T-3′ and reverse 5′-GCC GGT GTA GGT GTA GA-3′) and GAPDH (forward, 5′- GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′- GAAGATGGCGATGGGATTTC-3′).
Relative quantification was performed using the 2
−ΔΔCT method [
20]. Results were normalized to the level of the housekeeping gene,
GAPDH, and were expressed as relative quantification (RQ; 2
−ΔΔCt).
2.7. Statistical Analysis
All statistical analyses were performed using Prism version 7.0 (GraphPad Software, La Jolla, CA, USA). IC50 (concentration required to inhibit cell proliferation by 50%) values were calculated from the linear transformation of dose–response curves. IC50 values were calculated by fitting data into dose–response curves (inhibitor vs. response) by nonlinear regression with a variable slope. Comparisons between two groups were evaluated with two-tailed Student’s t-tests. Experimental data including more than 2 groups were analyzed using one-way or two-way ANOVA. The data were considered statistically significant at p < 0.05.
4. Discussion
Three-dimensional cultures of cancer cells enable better mimicking of physiological conditions than traditional monolayer 2D cultures and are becoming a valuable tool in cancer cell biology research and drug screening. During recent years, different strategies have been exploited to study the behavior of cancer cells in 3D milieus thanks to the employment of advanced biotechnologies, different kinds of scaffolds, and the most sophisticated culture approaches [
25]. According to the technology used, 3D cancer models include a variety of systems, such as scaffold-free systems, scaffold-based systems, hydrogel-based models, bioreactor-based models, microcarrier-based models, and cancer-on-a-chip [
26]. In addition, bioprinting has been recently proposed to develop 3D models that combine cells and biomaterials (bioink) to obtain tissue-like structures by means of a layer-by-layer deposition [
27]. In this study, we simultaneously printed OS cells (U-2 OS or its CDDP-resistant variant) together with a collagen bioink to develop collagen-based hydrogel constructs aimed at creating OS tumor models that better mimicked the features of the native microenvironment, and we studied the 3D tumor characteristics.
Cell survival rate is one of the key factors to consider while applying 3D cell printing technology in the construction of tissue-like models. Cells are subjected to mechanical forces during the 3D extrusion cell printing process, and it is well known that increased mechanical forces can cause cellular damage, reducing cell survival rate. Thus, one of the big challenges for bioprinting is to simultaneously print cells and the material (bioink) without affecting the cell viability. Here, we could confirm that the collagen-based hydrogels and the printing conditions used to model the bone extracellular matrix [
28] preserve the viability and metabolic activity of OS cells. The chemical composition, relative abundance, and spatial organization of ECM constituents confer each tissue type with unique physical and biochemical properties (e.g., the rigidity of the matrix or its porosity) that may affect cell behavior. Collagen is widely used for engineering hard tissues, such as bone, because of its strength and toughness [
29]. In addition, beyond mechanical properties, collagen fibers self-organize into 3D networks and can mimic the native matrix features, providing stimuli for cell adhesion, biodegradation sequences, and cues for supporting cell survival, growth, and migration [
30]. The polymerization properties of collagen are highly dependent on its concentration, temperature, pH, ionic forces, and level of cross-linking [
31]. Therefore, by properly tuning the polymerization parameters, we can change the mechanical properties of the scaffolds. In addition, in the design of the pore size, the physical hydrogel long-term swelling behavior and the need for higher porosity values should be considered to guarantee proper nutrient transport properties during culture conditions [
32].
Herein, we present a novel 3D system in which all these parameters have been optimized to reproduce the mechanical constrain sensed by cells in primary tumors, and that successfully sustained OS cell proliferation and migration.
Cell migration is crucial for metastasis, the most relevant clinical challenge for oncologists treating OS patients. Cell assays based on 2D cellular models, such as wound healing or scratch-based assays, are widely used for migration research, but these systems cannot faithfully recapitulate the molecular and biomechanical complexity of in vivo environments. Indeed, 2D models are not able to replicate some specific features of the tumor environment, such as the cell’s spatial confinement, or cell–cell and cell–matrix interactions that affect the proliferation or the response to migration stimuli. Several MMP family members are among the main contributors of the degradation of ECM during tumor cell invasion [
33].
In our experimental conditions, we found that either sensitive or resistant OS cells express higher levels of MMP-1, -9, and -13 in 3D compared to flat conditions. This discrepant behavior was particularly evident for the chemo-resistant U-2OS/CDDP 1 μg cells, which, in planar cultures, were less aggressive either in terms of invasion/migration or proliferation in comparison with parental cells, but that regained their aggressiveness when printed in collagen hydrogels.
Findings from the proliferation and migration assays, indeed, indicate that the collagen-based hydrogel is a suitable environment to preserve the tumor aggressiveness of CDDP-resistant OS cells. We can speculate that collagen may influence the proliferation of resistant cell more than that of sensitive ones because these cells have differential expression of collagen receptors on their surface. Cellular behavior is controlled by cell signal transduction pathways. Additionally, integrins (typical adhesion molecules in cancer cells), TGFβ receptors, and collagens closely associate with discoidin domain receptors (DDRs), a subfamily of tyrosine kinases that can be phosphorylated by collagen ligation, leading to the activation of AKT/PI3K signaling, mitogen-activated protein kinase (MAPK)/ERK signaling, and Rho family signaling [
34], which regulate various functions, including cell proliferation and migration. In addition, cancer cells can also influence the expression and production of collagens, and their remodeling. For example, proteolysis of collagen is thought to be crucial for the active movement of tumor cells, as it opens migratory tracks and reduces mechanical stress on the migrating cells. In addition, the remodeling of collagens may be essential for releasing soluble growth factors, such as the liberation of insulin-like growth factors that were reported to sustain OS cell proliferation [
30].
Thus, the highest expression of metalloproteinases that is induced by the 3D condition may explain the discrepancy in the positivity of Ki-67 that we observed for resistant cells between 3D and 2D cultures. Conversely, maintaining the cell-free construct in culture did not alter its structure over time. This indicates, on one side, the stability of the printed constructs, and, on the other side, gave evidence of the role exerted by MMPs in remodeling the collagen matrix after OS cell embedding.
Changes in the tumor microenvironment also affect drug sensitivity [
35]. In this study, we demonstrated that the 3D condition enhanced the expression of genes implicated in the repair of CDDP-induced DNA damages [
19,
36] before OS cells were exposed to any chemotherapeutic drug, and decreased sensitivity to chemotherapeutic agents. It is widely described that collagen plays an important role in therapy resistance [
34]. In addition, different collagen types may induce distinct treatment resistances. For example, in ER-positive cancer cells, COLI induced resistance to drugs, such as cisplatin and mitoxantrone, by activating β1 integrin followed by the FAK/PI3K/AKT pathway, whereas in triple-negative cancer cells, the MAPK pathway was found to be involved [
37]. COL11A1 induced chemoresistance and exerted anti-apoptotic effects in ovarian cancer cells by mediating the transcriptional activation of NF-κB to upregulate the Twist family [
38]. In our cellular model, the presence of collagen type I induced a strong expression of genes involved in DNA repair-related factors belonging to the nucleotide excision repair (NER) or base excision repair (BER) pathways and of the glutathione S-transferase P1 (GSTP1) either in sensitive or resistant cells. As proof-of-principle, we chose CDDP because it is a DNA-damaging agent currently used in the treatment of OS. Previous studies have clearly shown that overexpression of these genes negatively impact the OS cell responsiveness to CDDP-based treatments, as well as patient outcome [
19,
36]. Here, we show that the 3D hydrogel provided a better structural support than 2D conditions in fostering gene expression of DNA repair enzymes in sensitive and resistant OS cells. Overall, our data indicate that collagen-based hydrogels are suitable systems to study drug efficacy. These scaffolds provide reliable and reproducible results when used to measure the chemosensitivity of cells, and their use should be extended to other chemotherapeutics or targeted agents.