Critical Review on the Progress of Plastic Bioupcycling Technology as a Potential Solution for Sustainable Plastic Waste Management
Abstract
:1. Introduction
2. Recycling of Conventional Plastic Wastes
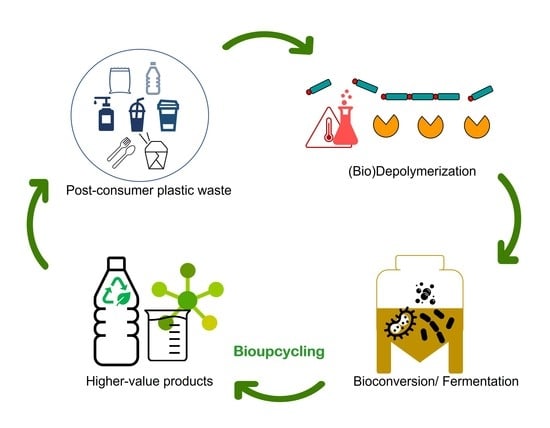
3. Bioupcycling
3.1. Bioupcycling of Polyethylene Terephthalate (PET)
| Depolymerization Strategy | Depolymerization Products Used as a Feedstock for Fermentation Step | Fermentation Strategy | Products from Fermentation | Titer | Productivity | Yield | Ref. |
|---|---|---|---|---|---|---|---|
| Hydrolytic pyrolysis at 450 °C | Solid product mixture (terephthalic acid (TA), oligomers, benzoic acid, and others) | Fermentation in shake flask containing 4.2 g/L of PET-derived sodium terephthalate and 67 mg/L of nitrogen at 30 °C for 48 h by Pseudomonas putida GO16 | medium chain length PHA (mclPHA) | 0.25 g/L | 8.4 mgPHA/L/h | 0.27 gPHA/gCDW | [63] |
| Hydrolytic pyrolysis at 450 °C | Solid product mixture (TA, oligomers, benzoic acid, and others) | Fermentation in shake flask containing 4.2 g/L of PET-derived sodium terephthalate and 67 mg/L of nitrogen at 30 °C for 48 h by P. putida GO19 | mclPHA | 0.25 g/L | 8.4 mgPHA/L/h | 0.23 gPHA/gCDW | [63] |
| Hydrolytic pyrolysis at 450 °C | Solid product mixture (TA, oligomers, benzoic acid, and others) | Fermentation in shake flask containing 4.2 g/L of PET-derived sodium terephthalate and 67 mg/L of nitrogen at 30 °C for 48 h by P. putida GO23 | mclPHA | 0.27 g/L | 4.4 mgPHA/L/h | 0.24 gPHA/gCDW | [63] |
| Pyrolysis | TA | Fed-batch fermentation in 19.5 L-stirred tank reactor with controlled pH of 6.9 and dissolved oxygen (DO) level above 40% at 30 °C for 48 h by P. putida GO16 supplied with TA as the sole growth and PHA substrate | mclPHA | 2.61 g/L | 0.05 g/L/h | 0.30 gPHA/gCDW | [66] |
| Pyrolysis | TA | Fed-batch fermentation in 19.5 L-stirred tank reactor with controlled pH of 6.9 and DO level above 40% at 30 °C for 48 h by P. putida GO16 supplied with waste glycerol (WG) as growth substrate and TA as PHA substrate | mclPHA | 5.22 g/L | 0.11 g/L/h | 0.36 gPHA/gCDW | [66] |
| Pyrolysis | TA | Fed-batch fermentation in 19.5 L-stirred tank reactor with controlled pH of 6.9 and DO level above 40% at 30 °C for 48 h by P. putida GO16 supplied with TA as the sole growth and PHA substrate | mclPHA | 5.30 g/L | 0.11 g/L/h | 0.35 gPHA/gCDW | [66] |
| Pyrolysis | TA | Fed-batch fermentation in 19.5 L-stirred tank reactor with controlled pH of 6.9 and DO level above 40% at 30 °C for 48 h by P. putida GO16 supplied with WG as growth and PHA substrate and TA as PHA substrate only | mclPHA | 4.98 g/L | 0.10 g/L/h | 0.35 gPHA/gCDW | [66] |
| Pyrolysis | TA | Fed-batch fermentation in 19.5 L-stirred tank reactor with controlled pH of 6.9 and DO level above 40% at 30 °C for 48 h by P. putida GO16 supplied with WG and TA as both growth and PHA substrates | mclPHA | 4.42 g/L | 0.09 g/L/h | 0.36 gPHA/gCDW | [66] |
| Enzymatic degradation by recombinant leaf-branch compost cutinase (LCC) | TA, ethylene glycol (EG), mono-(2-hydroxyethyl)terephthalic acid (MHET), di-(2-hydroxyethyl)terephthalic acid (BHET) | Fermentation in 5 L-stirred tank reactor with controlled pH of 7.0 and DO level above 20% at 30 °C for 28 h Pseudomonas umsongensis GO16 KS3 supplied with hydrolyzed PET at the amount to yield 40 mM of TA and EG and limited inorganic nutrient | mclPHA | 0.15 g/L | NA | 0.014 gPHA/gSubstrate | [16] |
| Enzymatic degradation by recombinant LCC | TA, EG, MHET, BHET | Fermentation in shake flask containing Delf medium with diluted (1:20) hydrolyzed PET (TA and EG concentration of 15–18 mM) at 30 °C for 24 h by P. umsongensis GO16 KS3 pSB01 | Hydroxyalkanoyloxy-alkanoate (HAA) | 35 mg/L | 5 mg/L/h | 0.01 gHAA/gTA | [16] |
| Microwave radiation for 50 min at 230 °C | TA | Bioconversion by metabolically engineered E. coli strain PCA-1 and HBH-2 to convert TA to intermediate protocatechuic acid (PCA), and then to gallic acid (GA), at 30 °C and 250 rpm for 24 h in 50 mM Tris buffer (pH 7.0) containing 2% (w/v) glycerol | GA | 2.7 mM | NA | 0.925 MGA/MTA | [67] |
| Microwave radiation for 50 min at 230 °C | TA | Bioconversion by metabolically engineered E. coli strain PG-1a to convert TA to intermediate PCA, GA, and then pyrogallol, at 30 °C and 250 rpm for 6 h in 50 mM Tris buffer (pH 7.0) containing 2% (w/v) glycerol | Pyrogallol | 1.1 mM | NA | 0.327 MPyrogallol/MTA | [67] |
| Microwave radiation for 50 min at 230 °C | TA | Bioconversion for 6 h by metabolically engineered E. coli strain CTL-1 and MA-1 to convert TA to intermediate catechol, and then to muconic acid (MA) | MA | 2.7 mM | NA | 0.854 MMA/MTA | [67] |
| Microwave radiation for 50 min at 230 °C | TA | Bioconversion using double-catalyst VA-2a system for 48 h by metabolically engineered E. coli strain PCA-1 and OMT-2His to convert TA to intermediate PCA and then to vanillic acid (VA), in 50 mM Tris buffer (pH 7.0) containing 10% (w/v) glycerol, 10 g/L yeast extract, 20 g/L peptone, and 2.5 mM L-methionine | VA | 1.4 mM | NA | 0.416 MVA/MTA | [67] |
| Microwave radiation for 50 min at 230 °C | EG | Bioconversion by Gluconobacter oxydans KCCM 40109 using 10.7 mM of EG from PET hydrolysate as a feedstock, at 30 °C and 220 rpm in shake flask at the working volume of 1 L | Glycolic acid (GLA) | NA | NA | 0.986 MGLA/MEG | [67] |
| - | EG (mock substrate to study upcycling of PET-derived monomer) | Fermentation in shake flask containing 10% (v/v) EG in 250 mM potassium phosphate buffer (pH 7.0) at 30 °C with gentle stirring and aeration at 1 VVM for 120 h by Pichia naganishii AKU 4267 | GLA | 105 g/L | NA | 0.880 MGLA/MEG | [70] |
| - | EG (mock substrate to study upcycling of PET-derived monomer) | Fermentation in shake flask containing 10% (v/v) EG in 250 mM potassium phosphate buffer (pH 7.0) at 30 °C with gentle stirring and aeration at 1 VVM for 120 h by Rhodotorula sp. 3Pr-126 | GLA | 110 g/L | NA | 0.922 MGLA/MEG | [70] |
| - | EG (mock substrate to study upcycling of PET-derived monomer) | Fermentation in shake flask containing 100 mM of EG in nitrogen limiting M9 medium (0.132 g/L of (NH4)2SO4) at 30 °C for more than 72 h by P. putida MFL185 (engineered strain that has the tac promoter inserted before the native glycolate oxidase operon and harbor overexpression) | mclPHA | NA | NA | 0.32 gPHA/gCDW and 0.06 gPHA/gEG | [71] |
| - | EG (mock substrate to study upcycling of PET-derived monomer) | Anaerobic fermentation of 50 mM EG at 30 °C by acetogenic bacterium Acetobacterium woodii | Acetate | 10.4 mM | 3.6 μmol/mg/h | NA | [72] |
| - | EG (mock substrate to study upcycling of PET-derived monomer) | Anaerobic fermentation of 50 mM EG at 30 °C by acetogenic bacterium A. woodii | Ethanol | 12.0 mM | 4.8 μmol/mg/h | NA | [72] |
| Enzymatic degradation by semi-purified LCC (pH 10.0) at 72 °C for 48 h | PET hydrolysate | Bioconversion using metabolically engineered E. coli RARE_pVanX to convert TA to intermediate protocatechuate (PC), and then to vanillin using optimized condition: M9-glucose supplemented with L-Met and nBuOH as a protein expression media, pH 5.5, room temperature for 24 h, in situ product removal (ISPR) by oleyl alcohol | Vanillin | 300–400 μM | NA | NA | [68] |
| - | TA (mock substrate to study upcycling of PET-derived monomer) | Bioconversion using metabolically engineered E. coli RARE_pVanX to convert TA to intermediate PC and then to vanillin using optimized condition: M9-glucose supplemented with L-Met and nBuOH as a protein expression media, pH 5.5, room temperature for 24 h, ISPR by oleyl alcohol | Vanillin | 789 μM | NA | 0.79 Mvanillin/MTA | [68] |
| Chemical glycolysis at 200 °C for 3 h | Mixture of BHET, MHET, and PET oligomers at 84.8, 7.7, and 8.7%, respectively | Enzymatic hydrolysis of the glycolyzed products (the mixture) into TA by Bacillus subtilis esterase (Bs2Est) (2 U/mL at 30 °C and 1000 rpm), following by producing catechol from PET hydrolysates using a catechol biosynthesis strain that was established using the combination of the TA degradation module and catechol biosynthesis module in E. coli (in 12 h) | Catechol | 5.97 mM | NA | 0.995 MCatechol/MTA | [69] |
| Chemocatalytic glycolysis: PET was glycolyzed with EG as a solvent (1:4 w/w) and catalyzed by 1% (w/w) titanium (IV) butoxide at 220 °C overnight | BHET | Fermentation in 3 L-bioreactor with batch culture in the first 4 h fed with 4-hydroxybenzoic acid to induce the β-ketoadipate pathway, followed by fed-batch culture using BHET as a carbon source (pulse adding at 9.1, 23.3, 32.8, 48.2 and 73.9 h). The sequential metabolic engineered Pseudomonas putida KT2440 (constitutive expression of native genes for EG utilization, expression of gene for TA catabolism, expression of PETase and MHETase for BHET hydrolysis, and gene deletion to enhance β-ketoadipic acid production) was used for bioconversion. | β-ketoadipic acid | 15.1 g/L | 0.16 g/L/h | 0.76 Mβ-ketoadipic acid/MBHET | [73] |
| Chemocatalytic glycolysis and enzymatic hydrolysis: PET was glycolyzed with EG as a solvent and catalyzed by betaine at 190 °C for 30–120 min, followed by enzymatic hydrolysis (PETase and MHETase) | TA | Whole cell bioconversion of TA (4.5 g/L) to protocatechuic acid by metabolically engineered E. coli PCA-1 was performed in shake flask at 30 °C and 250 rpm | PCA | 3.8 g/L | - | 0.904 MPCA/MTA | [74] |
| Chemocatalytic glycolysis and enzymatic hydrolysis: PET was glycolyzed with EG as a solvent and catalyzed by betaine at 190 °C for 30–120 min, followed by enzymatic hydrolysis (PETase and MHETase) | EG | Whole cell bioconversion of EG (30.6 g/L) to GLA by Gluconobacter oxydan KCCM 40109 was performed in shake flask at 30 °C and 200 rpm. | GLA | 31.4 g/L | - | 0.916 MGLA/MEG | [74] |
3.2. Bioupcycling of Polyurethanes (PU)
3.3. Bioupcycling of Polyolefins
3.3.1. Polyethylene (PE)
- 1.
- Terminal oxidation:
- 2.
- Bi-terminal oxidation:
- 3.
- Subterminal oxidation:
| Depolymerization Strategy | Depolymerization Products Used as a Feedstock for Fermentation Step | Fermentation Strategy | Products from Fermentation | Titer | Productivity | Yield | Ref. |
|---|---|---|---|---|---|---|---|
| Pyrolysis | PE hydrolysis wax (a mixture of hydrocarbons (C8–C32): 90% alkanes and 10% alkenes) | Fermentation in shake flask containing 0.05% (w/v) PE pyrolysis wax as a sole carbon source and 0.025% (w/v) of NH4Cl as a nitrogen source at 30 °C for 48 h by Pseudomonas aeruginosa GL-1 | PHA | 0.023 g/L | NA | 0.10 gPHA/gCDW | [120] |
| Pyrolysis | PE hydrolysis wax (a mixture of hydrocarbons (C8–C32): 90% alkanes and 10% alkenes) | Fermentation in shake flask containing 2% (w/v) PE pyrolysis wax as a sole carbon source and 0.019% (w/v) of NH4NO3 as a nitrogen source at 30 °C for 48 h by P. aeruginosa GL-1, in the presence of 0.05% (w/v) rhamnolipids | PHA | 0.074 g/L | NA | 0.19 gPHA/gCDW | [120] |
| Pyrolysis | PE hydrolysis wax (a mixture of hydrocarbons (C8–C32): 90% alkanes and 10% alkenes) | Fermentation in shake flask containing 2% (w/v) PE pyrolysis wax as a sole carbon source and 0.019% (w/v) of NH4NO3 as a nitrogen source at 30 °C for 48 h by P. aeruginosa PAO1, in the presence of 0.05% (w/v) rhamnolipids | PHA | 0.045 g/L | NA | 0.15 gPHA/gCDW | [120] |
| Oxidative degradation in a two-phase system (gas-liquid phase), after melting at 145 °C and using oxygen | Oxidized polyethylene wax (O-PEW) | Fermentation in shake flask containing 4 g/L melted O-PEW emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by Ralstonia eutropha H16 | PHA | 1.25 g/L | NA | 0.34 gPHA/gCDW | [121] |
| - | - | Fermentation in shake flask containing Ramsey’s media with 1% LDPE particles at 30 °C and 150 rpm for 21 d by Cuprividus necator H16 | short chain length PHA (sclPHA) | NA | NA | 0.0318 gPHA/gCDW | [123] |
| - | - | Fermentation in shake flask containing Ramsey’s media with 1% LDPE particles at 30 °C and 150 rpm for 21 d by Pseudomonas putida LS46 | mclPHA | NA | NA | 0.0054 gPHA/gCDW | [123] |
| - | - | Fermentation in shake flask containing Ramsey’s media with 1% LDPE particles at 30 °C and 150 rpm for 21 d by Acinetobacter pittii IRN19 | mclPHA | NA | NA | 0.0049 gPHA/gCDW | [123] |
| Pyrolysis | Non-oxidized PE wax (N-PEW) | Fermentation in shake flask containing 4 g/L melted N-PEW emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by Cupriavidus necator H16 | PHA | 0.46 g/L | NA | 0.32 gPHA/gCDW | [122] |
3.3.2. Polypropylene (PP)
3.4. Bioupcycling of Polystyrene (PS)
3.5. Bioupcycling of Polyvinyl Chloride (PVC)
3.6. Bioupcycling of Mixed Plastic Waste
4. Future Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OECD. Global Plastics Outlook: Economic Drivers, Environmental Impacts and Policy Options; OECD Publishing: Paris, France, 2022; ISBN 9789264584068. [Google Scholar]
- US EPA Plastics: Material-Specific Data. Available online: https://www.epa.gov/facts-and-figures-about-materials-waste-and-recycling/plastics-material-specific-data (accessed on 6 March 2022).
- PlasticsEurope The Facts 2021. Available online: https://plasticseurope.org/knowledge-hub/plastics-the-facts-2021/ (accessed on 16 February 2022).
- European Commission. A European Strategy for Plastics in a circular economy. Eur. Com. 2018. [Google Scholar]
- Hamilton, L.A.; Feit, S.; Muffett, C.; Kelso, M.; Rubright, S.M.; Bernhardt, C.; Schaeffer, E.; Moon, D.; Morris, J.; Labbé-Bellas, R. Plastic & Climate the Hidden Costs of a Plastic Planet; Center for International Environmental Law (CIEL): Washington, DC, USA, 2019. [Google Scholar]
- The Ocean Conference Factsheet: Marine Pollution. Available online: https://sustainabledevelopment.un.org/content/documents/Ocean_Factsheet_Pollution.pdf (accessed on 6 March 2022).
- Liebmann, B.; Köppel, S.; Reiberger, T.; Schwab, P. Assessment of Microplastic Concentrations in Human Stool-Preliminary Results of a Prospective Study. In Proceedings of the United European Gastroenterology (UEG) Week, Vienna, Austria, 20–24 October 2018; pp. 1–16. [Google Scholar]
- Sharma, S.; Chatterjee, S. Microplastic pollution, a threat to marine ecosystem and human health: A short review. Environ. Sci. Pollut. Res. 2017, 24, 21530–21547. [Google Scholar] [CrossRef] [PubMed]
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lebreton, L.; Egger, M.; Slat, B. A global mass budget for positively buoyant macroplastic debris in the ocean. Sci. Rep. 2019, 9, 12922. [Google Scholar] [CrossRef] [Green Version]
- European Commission. Circular Economy Action Plan. Available online: https://ec.europa.eu/environment/strategy/circular-economy-action-plan_en (accessed on 7 March 2022).
- Roux, M.; Varrone, C. Assessing the Economic Viability of the Plastic Biorefinery Concept and Its Contribution to a More Circular Plastic Sector. Polymer 2021, 13, 3883. [Google Scholar] [CrossRef]
- Lomwongsopon, P.; Varrone, C. Contribution of Fermentation Technology to Building Blocks for Renewable Plastics. Fermentation 2022, 8, 47. [Google Scholar] [CrossRef]
- Wierckx, N.; Prieto, M.A.; Pomposiello, P.; de Lorenzo, V.; O’Connor, K.; Blank, L.M. Plastic waste as a novel substrate for industrial biotechnology. Microb. Biotechnol. 2015, 8, 900–903. [Google Scholar] [CrossRef]
- Johnston, B.; Radecka, I.; Hill, D.; Chiellini, E.; Ilieva, V.; Sikorska, W.; Musioł, M.; Zięba, M.; Marek, A.; Keddie, D.; et al. The Microbial Production of Polyhydroxyalkanoates from Waste Polystyrene Fragments Attained Using Oxidative Degradation. Polymers 2018, 10, 957. [Google Scholar] [CrossRef] [Green Version]
- Tiso, T.; Narancic, T.; Wei, R.; Pollet, E.; Beagan, N.; Schröder, K.; Honak, A.; Jiang, M.; Kenny, S.T.; Wierckx, N.; et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 2021, 66, 167–178. [Google Scholar] [CrossRef]
- Narancic, T.; Salvador, M.; Hughes, G.M.; Beagan, N.; Abdulmutalib, U.; Kenny, S.T.; Wu, H.; Saccomanno, M.; Um, J.; O’Connor, K.E.; et al. Genome analysis of the metabolically versatile Pseudomonas umsongensis GO16: The genetic basis for PET monomer upcycling into polyhydroxyalkanoates. Microb. Biotechnol. 2021, 14, 2463–2480. [Google Scholar] [CrossRef]
- Tokiwa, Y.; Calabia, B.P.; Ugwu, C.U.; Aiba, S. Biodegradability of Plastics. Int. J. Mol. Sci. 2009, 10, 3722–3742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, D.; Su, F.; Liu, C.; Guo, Z. Research progress for plastic waste management and manufacture of value-added products. Adv. Compos. Hybrid Mater. 2020, 3, 443–461. [Google Scholar] [CrossRef]
- Babaremu, K.O.; Okoya, S.A.; Hughes, E.; Tijani, B.; Teidi, D.; Akpan, A.; Igwe, J.; Karera, S.; Oyinlola, M.; Akinlabi, E.T. Sustainable plastic waste management in a circular economy. Heliyon 2022, 8, e09984. [Google Scholar] [CrossRef] [PubMed]
- Idumah, C.I.; Nwuzor, I.C. Novel trends in plastic waste management. SN Appl. Sci. 2019, 1, 1402. [Google Scholar] [CrossRef] [Green Version]
- Ragaert, K.; Delva, L.; Geem, K.V. Mechanical and chemical recycling of solid plastic waste. Waste Manag. 2017, 69, 24–58. [Google Scholar] [CrossRef]
- Gala, A.; Guerrero, M.; Serra, J.M. Characterization of post-consumer plastic film waste from mixed MSW in Spain: A key point for the successful implementation of sustainable plastic waste management strategies. Waste Manag. 2020, 111, 22–33. [Google Scholar] [CrossRef]
- Hahladakis, J.N.; Velis, C.A.; Weber, R.; Iacovidou, E.; Purnell, P. An overview of chemical additives present in plastics: Migration, release, fate and environmental impact during their use, disposal and recycling. J. Hazard. Mater. 2018, 344, 179–199. [Google Scholar] [CrossRef]
- Hatti-Kaul, R.; Nilsson, L.J.; Zhang, B.; Rehnberg, N.; Lundmark, S. Designing Biobased Recyclable Polymers for Plastics. Trends Biotechnol. 2020, 38, 50–67. [Google Scholar] [CrossRef]
- Abu-Thabit, N.Y.; Pérez-Rivero, C.; Uwaezuoke, O.J.; Ngwuluka, N.C. From waste to wealth: Upcycling of plastic and lignocellulosic wastes to PHAs. J. Chem. Technol. Biotechnol. 2021, 97, 3217–3240. [Google Scholar] [CrossRef]
- Jamil, N.; Kumar, P.; Batool, R. Soil Microenvironment for Bioremediation and Polymer Production; John Wiley & Sons: Hoboken, NJ, USA, 2019; ISBN 9781119592129. [Google Scholar]
- Soni, V.K.; Singh, G.; Vijayan, B.K.; Chopra, A.; Kapur, G.S.; Ramakumar, S.S.V. Thermochemical Recycling of Waste Plastics by Pyrolysis: A Review. Energy Fuels 2021, 35, 12763–12808. [Google Scholar] [CrossRef]
- Thiounn, T.; Smith, R.C. Advances and approaches for chemical recycling of plastic waste. J. Polym. Sci. 2020, 58, 1347–1364. [Google Scholar] [CrossRef]
- Enthaler, S. Illustrating Plastic Production and End-of-Life Plastic Treatment with Interlocking Building Blocks. J. Chem. Educ. 2017, 94, 1746–1751. [Google Scholar] [CrossRef]
- Fox, J.A.; Stacey, N.T. Process targeting: An energy based comparison of waste plastic processing technologies. Energy 2019, 170, 273–283. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Vollmer, I.; Jenks, M.J.F.; Roelands, M.C.P.; White, R.J.; van Harmelen, T.; de Wild, P.; van der Laan, G.P.; Meirer, F.; Keurentjes, J.T.F.; Weckhuysen, B.M. Beyond Mechanical Recycling: Giving New Life to Plastic Waste. Angew. Chem. Int. Ed. 2020, 59, 15402–15423. [Google Scholar] [CrossRef] [Green Version]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.Y.; Choi, S.Y.; Lee, S.Y.; Kim, K.J. Rational Protein Engineering of Thermo-Stable PETase from Ideonella sakaiensis for Highly Efficient PET Degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Carniel, A.; Valoni, É.; Nicomedes, J.; Gomes, A.d.C.; de Castro, A.M. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Proc. Biochem. 2017, 59, 84–90. [Google Scholar] [CrossRef]
- Furukawa, M.; Kawakami, N.; Tomizawa, A.; Miyamoto, K. Efficient Degradation of Poly(ethylene terephthalate) with Thermobifida fusca Cutinase Exhibiting Improved Catalytic Activity Generated using Mutagenesis and Additive-based Approaches. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yao, C.; Xia, W.; Dou, M.; Du, Y.; Wu, J. Oxidative degradation of UV-irradiated polyethylene by laccase-mediator system. J. Hazard. Mater. 2022, 440, 129709. [Google Scholar] [CrossRef]
- Zhang, H.; Kong, D.; Wang, L.; Xia, W.; Yao, C.; Wu, J. Degradation of UV-pretreated polyolefins by latex clearing protein from Streptomyces sp. Strain K30. Sci. Total Environ. 2022, 806, 150779. [Google Scholar] [CrossRef]
- Sanluis-Verdes, A.; Colomer-Vidal, P.; Rodríguez-Ventura, F.; Bello-Villarino, M.; Spinola-Amilibia, M.; Ruiz-López, E.; Illanes-Vicioso, R.; Castroviejo, P.; Cigliano, R.A.; Montoya, M.; et al. Wax worm saliva and the enzymes therein are the key to polyethylene degradation by Galleria mellonella. bioRxiv 2022, 13, 5568. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.J.; Kim, M.N. Functional analysis of alkane hydroxylase system derived from Pseudomonas aeruginosa E7 for low molecular weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 2015, 103, 141–146. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.; Wu, W.M.; Zhao, J.; Song, Y.; Gao, L.; Yang, R.; Jiang, L. Biodegradation and Mineralization of Polystyrene by Plastic-Eating Mealworms: Part 1. Chemical and Physical Characterization and Isotopic Tests. Environ. Sci. Technol. 2015, 49, 12080–12086. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liu, H.; Gao, S.; Weng, Y.; Zhu, L. Enhanced extracellular production of IsPETase in Escherichia coli via engineering of the pelB signal peptide. J. Agric. Food Chem. 2021, 69, 2245–2252. [Google Scholar] [CrossRef]
- Rennison, A.; Winther, J.R.; Varrone, C. Rational Protein Engineering to Increase the Activity and Stability of IsPETase Using the PROSS Algorithm. Polymer 2021, 13, 3884. [Google Scholar] [CrossRef]
- Nakamura, A.; Kobayashi, N.; Koga, N.; Iino, R. Positive charge introduction on the surface of thermostabilized PET hydrolase facilitates PET binding and degradation. ACS Catal. 2021, 11, 8550–8564. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- Roth, C.; Wei, R.; Oeser, T.; Then, J.; Föllner, C.; Zimmermann, W.; Sträter, N. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 2014, 98, 7815–7823. [Google Scholar] [CrossRef]
- Singh, A.; Rorrer, N.A.; Nicholson, S.R.; Erickson, E.; DesVeaux, J.S.; Avelino, A.F.T.; Lamers, P.; Bhatt, A.; Zhang, Y.; Avery, G.; et al. Techno-economic, life-cycle, and socioeconomic impact analysis of enzymatic recycling of poly(ethylene terephthalate). Joule 2021, 5, 2479–2503. [Google Scholar] [CrossRef]
- Ellis, L.D.; Rorrer, N.A.; Sullivan, K.P.; Otto, M.; McGeehan, J.E.; Román-Leshkov, Y.; Wierckx, N.; Beckham, G.T. Chemical and biological catalysis for plastics recycling and upcycling. Nat. Catal. 2021, 4, 539–556. [Google Scholar] [CrossRef]
- Jenkins, S.; Quer, A.M.I.; Fonseca, C.; Varrone, C. Microbial Degradation of Plastics: New Plastic Degraders, Mixed Cultures and Engineering Strategies. In Soil Microenvironment for Bioremediation and Polymer Production; Jamil, N., Kumar, P., Batool, R., Eds.; Wiley Online Book: Hoboken, NJ, USA, 2019; pp. 215–238. ISBN 9781119592129. [Google Scholar]
- Arkatkar, A.; Arutchelvi, J.; Bhaduri, S.; Uppara, P.V.; Doble, M. Degradation of unpretreated and thermally pretreated polypropylene by soil consortia. Int. Biodeterior. Biodegrad. 2009, 63, 106–111. [Google Scholar] [CrossRef]
- Yoon, M.G.; Jeon, H.J.; Kim, M.N. Biodegradation of Polyethylene by a Soil Bacterium and AlkB Cloned Recombinant Cell. J. Bioremediat. Biodegrad. 2012, 3, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and Enzymatic Degradation of Synthetic Plastics. Front. Microbiol. 2020, 11, 2837. [Google Scholar] [CrossRef] [PubMed]
- Tiseo, I. PET Demand Worldwide 2030. Available online: https://www.statista.com/statistics/1128658/polyethylene-terephthalate-demand-worldwide/ (accessed on 6 October 2022).
- Zero Waste Europe New Report: PET is Far from Real Circularity. Available online: https://www.recycling-magazine.com/2022/02/16/new-report-pet-is-far-from-real-circularity/ (accessed on 16 February 2022).
- Hou, Q.; Zhen, M.; Qian, H.; Nie, Y.; Bai, X.; Xia, T.; Rehman, M.L.U.; Li, Q.; Ju, M. Upcycling and catalytic degradation of plastic wastes. Cell Rep. Phys. Sci. 2021, 2, 100514. [Google Scholar] [CrossRef]
- Chen, C.C.; Han, X.; Li, X.; Jiang, P.; Niu, D.; Ma, L.; Liu, W.; Li, S.; Qu, Y.; Hu, H.; et al. General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis. Nat. Catal. 2021, 4, 425–430. [Google Scholar] [CrossRef]
- Brott, S.; Pfaff, L.; Schuricht, J.; Schwarz, J.N.; Böttcher, D.; Badenhorst, C.P.S.; Wei, R.; Bornscheuer, U.T. Engineering and evaluation of thermostable IsPETase variants for PET degradation. Eng. Life Sci. 2022, 22, 192–203. [Google Scholar] [CrossRef]
- Cui, L.; Qiu, Y.; Liang, Y.; Du, C.; Dong, W.; Cheng, C.; He, B. Excretory expression of IsPETase in E. coli by an enhancer of signal peptides and enhanced PET hydrolysis. Int. J. Biol. Macromol. 2021, 188, 568–575. [Google Scholar] [CrossRef]
- Ferrario, V.; Todea, A.; Wolansky, L.; Piovesan, N.; Guarneri, A.; Ribitsch, D.; Guebitz, G.M.; Gardossi, L.; Pellis, A. Effect of Binding Modules Fused to Cutinase on the Enzymatic Synthesis of Polyesters. Catalyst 2022, 12, 303. [Google Scholar] [CrossRef]
- Knott, B.C.; Erickson, E.; Allen, M.D.; Gado, J.E.; Graham, R.; Kearns, F.L.; Pardo, I.; Topuzlu, E.; Anderson, J.J.; Austin, H.P.; et al. Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc. Natl. Acad. Sci. USA 2020, 117, 25476–25485. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Haugwitz, G.V.; Pfaff, L.; Mican, J.; Badenhorst, C.P.S.; Liu, W.; Weber, G.; Austin, H.P.; Bednar, D.; Damborsky, J.; et al. Mechanism-Based Design of Efficient PET Hydrolases. ACS Catal. 2022, 12, 3382–3396. [Google Scholar] [CrossRef] [PubMed]
- Kenny, S.T.; Runic, J.N.; Kaminsky, W.; Woods, T.; Babu, R.P.; Keely, C.M.; Blau, W.; O’Connor, K.E. Up-cycling of PET (Polyethylene Terephthalate) to the biodegradable plastic PHA (Polyhydroxyalkanoate). Environ. Sci. Technol. 2008, 42, 7696–7701. [Google Scholar] [CrossRef] [PubMed]
- Saratale, R.G.; Cho, S.K.; Saratale, G.D.; Kadam, A.A.; Ghodake, G.S.; Kumar, M.; Bharagava, R.N.; Kumar, G.; Kim, D.S.; Mulla, S.I.; et al. A comprehensive overview and recent advances on polyhydroxyalkanoates (PHA) production using various organic waste streams. Bioresour. Technol. 2021, 325, 124685. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Cho, I.J.; Lee, Y.; Kim, Y.J.; Kim, K.J.; Lee, S.Y. Microbial Polyhydroxyalkanoates and Nonnatural Polyesters. Adv. Mater. 2020, 32, 1907138. [Google Scholar] [CrossRef]
- Kenny, S.T.; Runic, J.N.; Kaminsky, W.; Woods, T.; Babu, R.P.; O’Connor, K.E. Development of a bioprocess to convert PET derived terephthalic acid and biodiesel derived glycerol to medium chain length polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 2012, 95, 623–633. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.K.; Cha, H.G.; Kang, M.J.; Lee, H.S.; Khang, T.U.; Yun, E.J.; Lee, D.H.; Song, B.K.; Park, S.J.; et al. Biological Valorization of Poly(ethylene terephthalate) Monomers for Upcycling Waste PET. ACS Sustain. Chem. Eng. 2019, 7, 19396–19406. [Google Scholar] [CrossRef]
- Sadler, J.C.; Wallace, S. Microbial synthesis of vanillin from waste poly(ethylene terephthalate). Green Chem. 2021, 23, 4665–4672. [Google Scholar] [CrossRef]
- Kim, H.T.; Ryu, M.H.; Jung, Y.J.; Lim, S.; Song, H.M.; Park, J.; Hwang, S.Y.; Lee, H.S.; Yeon, Y.J.; Sung, B.H.; et al. Chemo-Biological Upcycling of Poly(ethylene terephthalate) to Multifunctional Coating Materials. ChemSusChem 2021, 14, 4251–4259. [Google Scholar] [CrossRef]
- Kataoka, M.; Sasaki, M.; Hidalgo, A.R.G.D.; Nakano, M.; Shimizu, S. Glycolic acid production using ethylene glycol-oxidizing microorganisms. Biosci. Biotechnol. Biochem. 2001, 65, 2265–2270. [Google Scholar] [CrossRef] [Green Version]
- Franden, M.A.; Jayakody, L.N.; Li, W.J.; Wagner, N.J.; Cleveland, N.S.; Michener, W.E.; Hauer, B.; Blank, L.M.; Wierckx, N.; Klebensberger, J.; et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab. Eng. 2018, 48, 197–207. [Google Scholar] [CrossRef]
- Trifunović, D.; Schuchmann, K.; Müller, V. Ethylene glycol metabolism in the acetogen Acetobacterium woodii. J. Bacteriol. 2016, 198, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Werner, A.Z.; Clare, R.; Mand, T.D.; Pardo, I.; Ramirez, K.J.; Haugen, S.J.; Bratti, F.; Dexter, G.N.; Elmore, J.R.; Huenemann, J.D.; et al. Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 2021, 67, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Han, D.O.; Shim, K.I.; Kim, J.K.; Pelton, J.G.; Ryu, M.H.; Joo, J.C.; Han, J.W.; Kim, H.T.; Kim, K.H. One-Pot Chemo-bioprocess of PET Depolymerization and Recycling Enabled by a Biocompatible Catalyst, Betaine. ACS Catal. 2021, 11, 3996–4008. [Google Scholar] [CrossRef]
- Utomo, R.N.C.; Li, W.J.; Tiso, T.; Eberlein, C.; Doeker, M.; Heipieper, H.J.; Jupke, A.; Wierckx, N.; Blank, L.M. Defined Microbial Mixed Culture for Utilization of Polyurethane Monomers. ACS Sustain. Chem. Eng. 2020, 8, 17466–17474. [Google Scholar] [CrossRef]
- Mahajan, N.; Gupta, P. New insights into the microbial degradation of polyurethanes. RSC Adv. 2015, 5, 41839–41854. [Google Scholar] [CrossRef]
- Bisceglie, F.D.; Quartinello, F.; Vielnascher, R.; Guebitz, G.M.; Pellis, A. Cutinase-Catalyzed Polyester-Polyurethane Degradation: Elucidation of the Hydrolysis Mechanism. Polymer 2022, 14, 411. [Google Scholar] [CrossRef]
- Liang, C.; Gracida-Alvarez, U.R.; Gallant, E.T.; Gillis, P.A.; Marques, Y.A.; Abramo, G.P.; Hawkins, T.R.; Dunn, J.B. Material Flows of Polyurethane in the United States. Environ. Sci. Technol. 2021, 55, 14215–14224. [Google Scholar] [CrossRef]
- Liu, J.; He, J.; Xue, R.; Xu, B.; Qian, X.; Xin, F.; Blank, L.M.; Zhou, J.; Wei, R.; Dong, W.; et al. Biodegradation and up-cycling of polyurethanes: Progress, challenges, and prospects. Biotechnol. Adv. 2021, 48, 107730. [Google Scholar] [CrossRef]
- Europur The Netherlands: A World-Leader in Mattress Recycling. Available online: https://www.europur.org/news-events/item/70-the-netherlands-a-world-leader-in-mattress-recycling (accessed on 18 February 2022).
- Robinson, S. Recycling Polyols Back into Flexible Foam. Available online: https://www.utech-polyurethane.com/node/807346/printable/print (accessed on 18 February 2022).
- Cregut, M.; Bedas, M.; Durand, M.J.; Thouand, G. New insights into polyurethane biodegradation and realistic prospects for the development of a sustainable waste recycling process. Biotechnol. Adv. 2013, 31, 1634–1647. [Google Scholar] [CrossRef]
- Jin, X.; Dong, J.; Guo, X.; Ding, M.; Bao, R.; Luo, Y. Current advances in polyurethane biodegradation. Polym. Int. 2022, 71, 1384–1392. [Google Scholar] [CrossRef]
- Gewert, B.; Plassmann, M.M.; Macleod, M. Pathways for degradation of plastic polymers floating in the marine environment. Environ. Sci. Proc. Impacts 2015, 17, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Shah, Z.; Krumholz, L.; Aktas, D.F.; Hasan, F.; Khattak, M.; Shah, A.A. Degradation of polyester polyurethane by a newly isolated soil bacterium, Bacillus subtilis strain MZA-75. Biodegradation 2013, 24, 865–877. [Google Scholar] [CrossRef]
- Shah, Z.; Hasan, F.; Krumholz, L.; Atkas, D.; Shah, A.A. Degradation of polyester polyurethane by newly isolated Pseudomonas aeruginosa strain MZA-85 and analysis of degradation products by GC-MS. Int. Biodeterior. Biodegrad. 2013, 77, 114–122. [Google Scholar] [CrossRef]
- Shah, Z.; Gulzar, M.; Hasan, F.; Shah, A.A. Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polym. Degrad. Stab. 2016, 134, 349–356. [Google Scholar] [CrossRef]
- Vargas-Suárez, M.; Fernández-Cruz, V.; Loza-Tavera, H. Biodegradation of polyacrylic and polyester polyurethane coatings by enriched microbial communities. Appl. Microbiol. Biotechnol. 2019, 103, 3225–3236. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Nadir, S.; Shah, Z.U.; Shah, A.A.; Karunarathna, S.C.; Xu, J.; Khan, A.; Munir, S.; Hasan, F. Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environ. Pollut. 2017, 225, 469–480. [Google Scholar] [CrossRef]
- Magnin, A.; Hoornaert, L.; Pollet, E.; Laurichesse, S.; Phalip, V.; Avérous, L. Isolation and characterization of different promising fungi for biological waste management of polyurethanes. Microb. Biotechnol. 2019, 12, 544–555. [Google Scholar] [CrossRef]
- Álvarez-Barragán, J.; Domínguez-Malfavón, L.; Vargas-Suárez, M.; González-Hernández, R.; Aguilar-Osorio, G.; Loza-Tavera, H. Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 2016, 82, 5225–5235. [Google Scholar] [CrossRef] [Green Version]
- Zafar, U.; Nzeram, P.; Langarica-Fuentes, A.; Houlden, A.; Heyworth, A.; Saiani, A.; Robson, G.D. Biodegradation of polyester polyurethane during commercial composting and analysis of associated fungal communities. Bioresour. Technol. 2014, 158, 374–377. [Google Scholar] [CrossRef]
- Magnin, A.; Pollet, E.; Perrin, R.; Ullmann, C.; Persillon, C.; Phalip, V.; Avérous, L. Enzymatic recycling of thermoplastic polyurethanes: Synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag. 2019, 85, 141–150. [Google Scholar] [CrossRef]
- Bauer, A.S.; Tacker, M.; Uysal-Unalan, I.; Cruz, R.M.S.; Varzakas, T.; Krauter, V. Recyclability and Redesign Challenges in Multilayer Flexible Food Packaging—A Review. Foods 2021, 10, 2702. [Google Scholar] [CrossRef] [PubMed]
- Chamas, A.; Moon, H.; Zheng, J.; Qiu, Y.; Tabassum, T.; Jang, J.H.; Abu-Omar, M.; Scott, S.L.; Suh, S. Degradation Rates of Plastics in the Environment. ACS Sustain. Chem. Eng. 2020, 8, 3494–3511. [Google Scholar] [CrossRef] [Green Version]
- Ammala, A.; Bateman, S.; Dean, K.; Petinakis, E.; Sangwan, P.; Wong, S.; Yuan, Q.; Yu, L.; Patrick, C.; Leong, K.H. An overview of degradable and biodegradable polyolefins. Prog. Polym. Sci. 2011, 36, 1015–1049. [Google Scholar] [CrossRef]
- Albertsson, A.C.; Andersson, S.O.; Karlsson, S. The mechanism of biodegradation of polyethylene. Polym. Degrad. Stab. 1987, 18, 73–87. [Google Scholar] [CrossRef]
- Mukherjee, S.; Kundu, P.P. Alkaline fungal degradation of oxidized polyethylene in black liquor: Studies on the effect of lignin peroxidases and manganese peroxidases. J. Appl. Polym. Sci. 2014, 131, 8982–8990. [Google Scholar] [CrossRef]
- Sheik, S.; Chandrashekar, K.R.; Swaroop, K.; Somashekarappa, H.M. Biodegradation of gamma irradiated low density polyethylene and polypropylene by endophytic fungi. Int. Biodeterior. Biodegrad. 2015, 105, 21–29. [Google Scholar] [CrossRef]
- Pometto, A.L.; Lee, B.; Johnson, K.E. Production of an extracellular polyethylene-degrading enzyme (s) by Streptomyces species. Appl. Environ. Microbiol. 1992, 58, 731–733. [Google Scholar] [CrossRef] [Green Version]
- Iiyoshi, Y.; Tsutsumi, Y.; Nishida, T. Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. J. Wood Sci. 1998, 44, 222–229. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, Z.; Ma, X.; Liang, G.; Wang, J. Novel surface modification of high-density polyethylene films by using enzymatic catalysis. J. Appl. Polym. Sci. 2004, 91, 3673–3678. [Google Scholar] [CrossRef]
- Fujisawa, M. Degradation of polyethylene and nylon-66 by the laccase-mediator system. J. Polym. Environ. 2001, 9, 103–108. [Google Scholar] [CrossRef]
- Santo, M.; Weitsman, R.; Sivan, A. The role of the copper-binding enzyme—laccase—in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. Int. Biodeterior. Biodegrad. 2013, 84, 204–210. [Google Scholar] [CrossRef]
- Li, H.; Wang, X.L.; Mu, B.Z.; Gu, J.D.; Liu, Y.D.; Lin, K.F.; Lu, S.G.; Lu, Q.; Li, B.Z.; Li, Y.Y.; et al. Molecular detection, quantification and distribution of alkane-degrading bacteria in production water from low temperature oilfields. Int. Biodeterior. Biodegrad. 2013, 76, 49–57. [Google Scholar] [CrossRef]
- Yeom, S.J.; Le, T.K.; Yun, C.H. P450-driven plastic-degrading synthetic bacteria. Trends Biotechnol. 2022, 40, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.J.; Kim, M.N. Comparison of the functional characterization between alkane monooxygenases for low-molecular-weight polyethylene biodegradation. Int. Biodeterior. Biodegrad. 2016, 114, 202–208. [Google Scholar] [CrossRef]
- Watanabe, M.; Kawai, F.; Shibata, M.; Yokoyama, S.; Sudate, Y. Computational method for analysis of polyethylene biodegradation. J. Comput. Appl. Math. 2003, 161, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Montazer, Z.; Najafi, M.B.H.; Levin, D.B. Microbial degradation of low-density polyethylene and synthesis of polyhydroxyalkanoate polymers. Can. J. Microbiol. 2019, 65, 224–234. [Google Scholar] [CrossRef]
- Hammerer, L.; Winkler, C.K.; Kroutil, W. Regioselective Biocatalytic Hydroxylation of Fatty Acids by Cytochrome P450s. Catal. Lett. 2017, 148, 787–812. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Pedersen, J.N.; Eser, B.E.; Guo, Z. Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol. Adv. 2022, 60, 107991. [Google Scholar] [CrossRef]
- Peixoto, J.; Vizzotto, C.; Ramos, A.; Alves, G.; Steindorff, A.; Krüger, R. The role of nitrogen metabolism on polyethylene biodegradation. J. Hazard. Mater. 2022, 432, 128682. [Google Scholar] [CrossRef]
- Danso, D.; Chow, J.; Streita, W.R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019, 85, 2. [Google Scholar] [CrossRef] [Green Version]
- Raut, S.; Raut, S.; Sharma, M.; Srivastav, C.; Adhikari, B.; Sen, S.K. Enhancing Degradation of Low Density Polyethylene Films by Curvularia lunata SG1 Using Particle Swarm Optimization Strategy. Indian J. Microbiol. 2015, 55, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Mierzwa-Hersztek, M.; Gondek, K.; Kopeć, M. Degradation of Polyethylene and Biocomponent-Derived Polymer Materials: An Overview. J. Polym. Environ. 2019, 27, 600–611. [Google Scholar] [CrossRef] [Green Version]
- Restrepo-Flórez, J.M.; Bassi, A.; Thompson, M.R. Microbial degradation and deterioration of polyethylene-A review. Int. Biodeterior. Biodegrad. 2014, 88, 83–90. [Google Scholar] [CrossRef]
- Sohn, Y.J.; Kim, H.T.; Baritugo, K.A.; Jo, S.Y.; Song, H.M.; Park, S.Y.; Park, S.K.; Pyo, J.; Cha, H.G.; Kim, H.; et al. Recent Advances in Sustainable Plastic Upcycling and Biopolymers. Biotechnol. J. 2020, 15, 1900489. [Google Scholar] [CrossRef]
- Taghavi, N.; Singhal, N.; Zhuang, W.Q.; Baroutian, S. Degradation of plastic waste using stimulated and naturally occurring microbial strains. Chemosphere 2021, 263, 127975. [Google Scholar] [CrossRef] [PubMed]
- Koivuranta, K.; Andberg, M.B.; Nygren, H.; Castillo, S. Enzymes, Micro-Organisms and Uses Thereof, and a Method of Degrading Hydrocarbon Chains. International Patent No. WO2022/175592, 25 August 2022. [Google Scholar]
- Guzik, M.W.; Kenny, S.T.; Duane, G.F.; Casey, E.; Woods, T.; Babu, R.P.; Nikodinovic-Runic, J.; Murray, M.; O’Connor, K.E. Conversion of post consumer polyethylene to the biodegradable polymer polyhydroxyalkanoate. Appl. Microbiol. Biotechnol. 2014, 98, 4223–4232. [Google Scholar] [CrossRef]
- Radecka, I.; Irorere, V.; Jiang, G.; Hill, D.; Williams, C.; Adamus, G.; Kwiecień, M.; Marek, A.A.; Zawadiak, J.; Johnston, B.; et al. Oxidized polyethylene wax as a potential carbon source for PHA production. Materials 2016, 9, 367. [Google Scholar] [CrossRef] [Green Version]
- Johnston, B.; Jiang, G.; Hill, D.; Adamus, G.; Kwiecień, I.; Zięba, M.; Sikorska, W.; Green, M.; Kowalczuk, M.; Radecka, I. The Molecular Level Characterization of Biodegradable Polymers Originated from Polyethylene Using Non-Oxygenated Polyethylene Wax as a Carbon Source for Polyhydroxyalkanoate Production. Bioengineering 2017, 4, 73. [Google Scholar] [CrossRef] [Green Version]
- Montazer, Z.; Habibi-Najafi, M.B.; Mohebbi, M.; Oromiehei, A. Microbial Degradation of UV-Pretreated Low-Density Polyethylene Films by Novel Polyethylene-Degrading Bacteria Isolated from Plastic-Dump Soil. J. Polym. Environ. 2018, 26, 3613–3625. [Google Scholar] [CrossRef]
- Jeon, H.J.; Kim, M.N. Isolation of mesophilic bacterium for biodegradation of polypropylene. Int. Biodeterior. Biodegrad. 2016, 115, 244–249. [Google Scholar] [CrossRef]
- Skariyachan, S.; Taskeen, N.; Kishore, A.P.; Krishna, B.V.; Naidu, G. Novel consortia of enterobacter and pseudomonas formulated from cow dung exhibited enhanced biodegradation of polyethylene and polypropylene. J. Environ. Manag. 2021, 284, 112030. [Google Scholar] [CrossRef]
- Skariyachan, S.; Patil, A.A.; Shankar, A.; Manjunath, M.; Bachappanavar, N.; Kiran, S. Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polym. Degrad. Stab. 2018, 149, 52–68. [Google Scholar] [CrossRef]
- Yang, S.S.; Ding, M.Q.; He, L.; Zhang, C.H.; Li, Q.X.; Xing, D.F.; Cao, G.L.; Zhao, L.; Ding, J.; Ren, N.Q.; et al. Biodegradation of polypropylene by yellow mealworms (Tenebrio molitor) and superworms (Zophobas atratus) via gut-microbe-dependent depolymerization. Sci. Total Environ. 2020, 756, 144087. [Google Scholar] [CrossRef] [PubMed]
- Muenmee, S.; Chiemchaisri, W.; Chiemchaisri, C. Enhancement of biodegradation of plastic wastes via methane oxidation in semi-aerobic landfill. Int. Biodeterior. Biodegrad. 2016, 113, 244–255. [Google Scholar] [CrossRef]
- Johnston, B.; Radecka, I.; Chiellini, E.; Barsi, D.; Ilieva, V.I.; Sikorska, W.; Musiol, M.; Ziȩba, M.; Chaber, P.; Marek, A.A.; et al. Mass spectrometry reveals molecular structure of polyhydroxyalkanoates attained by bioconversion of oxidized polypropylene waste fragments. Polymers 2019, 11, 1580. [Google Scholar] [CrossRef] [Green Version]
- Mihreteab, M.; Stubblefield, B.A.; Gilbert, E.S. Microbial bioconversion of thermally depolymerized polypropylene by Yarrowia lipolytica for fatty acid production. Appl. Microbiol. Biotechnol. 2019, 103, 7729–7740. [Google Scholar] [CrossRef]
- Mihreteab, M.; Stubblefield, B.A.; Gilbert, E.S. Enhancing polypropylene bioconversion and lipogenesis by Yarrowia lipolytica using a chemical/biological hybrid process. J. Biotechnol. 2021, 332, 94–102. [Google Scholar] [CrossRef]
- Savoldelli, J.; Tomback, D.; Savoldelli, H. Breaking down polystyrene through the application of a two-step thermal degradation and bacterial method to produce usable byproducts. Waste Manag. 2017, 60, 123–126. [Google Scholar] [CrossRef]
- Farrelly, T.A.; Shaw, I.C. Polystyrene as Hazardous Household Waste. In Household Hazardous Waste Management; InTechOpen: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.R.; Lee, H.M.; Yu, H.C.; Jeon, E.; Lee, S.; Li, J.; Kim, D.H. Biodegradation of Polystyrene by Pseudomonas sp. Isolated from the Gut of Superworms (Larvae of Zophobas atratus). Environ. Sci. Technol. 2020, 54, 6987–6996. [Google Scholar] [CrossRef]
- Ho, B.T.; Roberts, T.K.; Lucas, S. An overview on biodegradation of polystyrene and modified polystyrene: The microbial approach. Crit. Rev. Biotechnol. 2018, 38, 308–320. [Google Scholar] [CrossRef]
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ. Pollut. 2017, 231, 1552–1559. [Google Scholar] [CrossRef] [PubMed]
- Syranidou, E.; Karkanorachaki, K.; Amorotti, F.; Franchini, M.; Repouskou, E.; Kaliva, M.; Vamvakaki, M.; Kolvenbach, B.; Fava, F.; Corvini, P.F.-X.; et al. Biodegradation of weathered polystyrene films in seawater microcosms. Sci. Rep. 2017, 7, 17991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Wang, J.; Xia, M. Biodegradation and mineralization of polystyrene by plastic-eating superworms Zophobas atratus. Sci. Total Environ. 2020, 708, 135233. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xin, X.; Shi, X.; Zhang, Y. A polystyrene-degrading Acinetobacter bacterium isolated from the larvae of Tribolium castaneum. Sci. Total Environ. 2020, 726, 138564. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.G.; Goff, M.; Donner, M.; Kaminsky, W.; O’Connor, K.E. A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ. Sci. Technol. 2006, 40, 2433–2437. [Google Scholar] [CrossRef]
- Goff, M.; Ward, P.G.; O’Connor, K.E. Improvement of the conversion of polystyrene to polyhydroxyalkanoate through the manipulation of the microbial aspect of the process: A nitrogen feeding strategy for bacterial cells in a stirred tank reactor. J. Biotechnol. 2007, 132, 283–286. [Google Scholar] [CrossRef]
- Ward, P.G.; Roo, G.D.; O’Connor, K.E. Accumulation of polyhydroxyalkanoate from styrene and phenylacetic acid by Pseudomonas putida CA-3. Appl. Environ. Microbiol. 2005, 71, 2046–2052. [Google Scholar] [CrossRef] [Green Version]
- Nikodinovic-Runic, J.; Casey, E.; Duane, G.F.; Mitic, D.; Hume, A.R.; Kenny, S.T.; O’Connor, K.E. Process analysis of the conversion of styrene to biomass and medium chain length polyhydroxyalkanoate in a two-phase bioreactor. Biotechnol. Bioeng. 2011, 108, 2447–2455. [Google Scholar] [CrossRef]
- Ru, J.; Huo, Y.; Yang, Y. Microbial Degradation and Valorization of Plastic Wastes. Front. Microbiol. 2020, 11, 442. [Google Scholar] [CrossRef] [Green Version]
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Polyvinyl chloride biodegradation by Pseudomonas citronellolis and Bacillus flexus. N. Biotechnol. 2019, 52, 35–41. [Google Scholar] [CrossRef]
- Peng, B.Y.; Chen, Z.; Chen, J.; Yu, H.; Zhou, X.; Criddle, C.S.; Wu, W.M.; Zhang, Y. Biodegradation of Polyvinyl Chloride (PVC) in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae. Environ. Int. 2020, 145, 106106. [Google Scholar] [CrossRef] [PubMed]
- Giacomucci, L.; Raddadi, N.; Soccio, M.; Lotti, N.; Fava, F. Biodegradation of polyvinyl chloride plastic films by enriched anaerobic marine consortia. Mar. Environ. Res. 2020, 158, 104949. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.I.; Ahmed, S.; Robson, G.; Javed, I.; Ali, N.; Atiq, N.; Hameed, A. Isolation and molecular characterization of polyvinyl chloride (PVC) plastic degrading fungal isolates. J. Basic Microbiol. 2014, 54, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; He, P.J.; Peng, W.; Yi, S.X.; Lü, F.; Shao, L.M.; Zhang, H. Upcycling waste polyvinyl chloride: One-pot synthesis of valuable carbon materials and pipeline-quality syngas via pyrolysis in a closed reactor. J. Hazard. Mater. 2022, 427, 128210. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Chen, D.; Yin, L.; Wang, Z.; Zhao, L.; Wang, J.Y. High efficiency chlorine removal from polyvinyl chloride (PVC) pyrolysis with a gas–liquid fluidized bed reactor. Waste Manag. 2014, 34, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Ballerstedt, H.; Tiso, T.; Wierckx, N.; Wei, R.; Averous, L.; Bornscheuer, U.; O’Connor, K.; Floehr, T.; Jupke, A.; Klankermayer, J.; et al. MIXed plastics biodegradation and UPcycling using microbial communities: EU Horizon 2020 project MIX-UP started January 2020. Environ. Sci. Eur. 2021, 33, 99. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.P.; Werner, Z.A.; Ramirez, K.J.; Ellis, L.E.; Bussard, J.R.; Black, B.A.; Brander, D.G.; Bratti, F.; Buss, B.L.; Dong, X.; et al. Mixed plastic waste valorization through tandem chemical oxidation and biological funneling. Science 2022, 378, 207–211. [Google Scholar] [CrossRef]
- Edwards, S.; León-zayas, R.; Ditter, R.; Laster, H.; Sheehan, G.; Anderson, O.; Beattie, T.; Mellies, J.L. Microbial Consortia and Mixed Plastic Waste: Pangenomic Analysis Reveals Potential for Degradation of Multiple Plastic Types via Previously Identified PET Degrading Bacteria. Int. J. Mol. Sci. 2022, 23, 5612. [Google Scholar] [CrossRef]
- European Comission. Waste Framework Directive. Available online: https://environment.ec.europa.eu/topics/waste-and-recycling/waste-framework-directive_en (accessed on 28 September 2022).
- Sheel, A.; Pant, D. Chemical Depolymerization of PET Bottles via Glycolysis. Recycl. Polyethyl. Terephthalate Bottles 2019, 1, 61–84. [Google Scholar] [CrossRef]
- Sinha, V.; Patel, M.R.; Patel, J.V. Pet waste management by chemical recycling: A review. J. Polym. Environ. 2010, 18, 8–25. [Google Scholar] [CrossRef]
- Gan, Z.; Zhang, H. PMBD: A Comprehensive Plastics Microbial Biodegradation Database. Database 2019, 2019, baz119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Wang, D.; Wei, N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022, 40, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Kremser, K.; Gerl, P.; Borrás, A.B.; Espinosa, D.R.; Martínez, B.M.; Guebitz, G.M.; Pellis, A. Bioleaching/enzyme-based recycling of aluminium and polyethylene from beverage cartons packaging waste. Resour. Conserv. Recycl. 2022, 185, 106444. [Google Scholar] [CrossRef]





| Depolymerization Strategy | Depolymerization Products Used as a Feedstock for Fermentation Step | Fermentation Strategy | Products from Fermentation | Titer | Productivity | Yield | Ref. |
|---|---|---|---|---|---|---|---|
| Enzymatic degradation of polycaprolactone polyol-based PU by esterase (E3576) in 0.1 M phosphate buffer (pH 7.0). The enzyme solution was replaced every 3–4 d to overcome a loss of enzymatic activity. | 6-hydroxycaproic acid (1 g/L) | - | - | - | - | - | [93] |
| - | Adipic acid (AA) (mock substrate to study upcycling of PU-derived monomer) | Bioconversion (at 30 °C and 200 rpm for 135 h) using metabolically engineered P. putida KT2440 A12.1p pPS05 to convert AA into HAA and then to rhamnolipid | Rhamnolipid | 0.02 g/L | NA | 0.014 gRhamnolipid/gSubstrate | [75] |
| - | 1,4-Butanediol (BDO) (mock substrate to study upcycling of PU-derived monomer) | Bioconversion (at 30 °C and 200 rpm for 135 h) using metabolically engineered P. putida KT2440 B10.1 pPR05 to convert BDO into HAA and then to rhamnolipid | Rhamnolipid | 0.13 g/L | NA | 0.088 gRhamnolipid/gSubstrate | [75] |
| - | EG (mock substrate to study upcycling of PU-derived monomer) | Bioconversion (at 30 °C and 200 rpm for 135 h) using metabolically engineered P. putida KT2440 ∆gclR ∆PP_2046 ∆PP_2662::14d to convert EG into HAA and then to rhamnolipid | Rhamnolipid | 0.07 g/L | NA | 0.038 gRhamnolipid/gSubstrate | [75] |
| - | AA + BDO + EG (mock hydrolysate to study upcycling of PU-derived monomers) | Bioconversion (at 30 °C and 200 rpm for 210 h) using mixed culture of three metabolically engineered P. putida KT2440 to convert the mock hydrolysate into HAA and then to rhamnolipid | Rhamnolipid | 0.1 g/L | NA | 0.008 gRhamnolipid/gSubstrate | [75] |
| AA + BDO + EG + 2,4-toluenediamine (TDA) (mock hydrolysate to study upcycling of PU-derived monomers) | Bioconversion (at 30 °C and 200 rpm for 210 h) using mixed culture of three metabolically engineered P. putida KT2440 to convert the mock hydrolysate into HAA and then to rhamnolipid without extraction of TDA | Rhamnolipid | 0.02 g/L | NA | 0.002 gRhamnolipid/gSubstrate | [75] | |
| AA + BDO + EG + TDA (mock hydrolysate to study upcycling of PU-derived monomers) | Bioconversion (at 30 °C and 200 rpm for 210 h) using mixed culture of three metabolically engineered P. putida KT2440 to convert the mock hydrolysate into HAA and then to rhamnolipid with extraction of TDA at pH 3.5 | Rhamnolipid | 0.07 g/L | NA | 0.005 gRhamnolipid/gSubstrate | [75] |
| Depolymerization Strategy | Depolymerization Products Used as a Feedstock for Fermentation Step | Fermentation Strategy | Products from Fermentation | Titer | Productivity | Yield | Ref. |
|---|---|---|---|---|---|---|---|
| Pro-degradation at 180 °C with 1% (w/w) cobalt stearate as pro-oxidant/pro-degradant additive | Oxidatively pro-degraded PP | Fermentation in shake flask containing 2 g/L oxidatively pro-degraded PP emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by C. necator H16 | PHA | 0.58 g/L | NA | 0.26 gPHA/gCDW | [129] |
| Oxidatively pro-degraded PP was subjected to oxidative degradation in a two-phase system (gas-liquid phase), after melting at 60–80 °C and using oxygen-ozone mixture | Thermal oxidized PP | Fermentation in shake flask containing 2 g/L thermal-oxidized PP emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by C. necator H16 | PHA | 1.36 g/L | NA | 0.42 gPHA/gCDW | [129] |
| Pyrolysis at 540 °C | Pyrolysis oil contained branched chain fatty alcohols (51%) and alkenes (25%) | Fermentation in shake flask containing OP4 medium (15 g/L pyrolysis oil, 5.4 g/L Tween 80, 4.5 g/L oleic acid, 1.25 g/L (NH4)2SO4, 2.5 g/L KH2PO4, and 0.830 g/L MgSO4·7H2O) at 30 °C for 312 h by Yarrowia lipolytica strain 78-003 | Fatty acids with C18 compounds (oleic acid, linoleic acid, and stearic acid) as dominant products, followed by C16 compounds (palmitic and palmitoleic acids). | 492 mg/L | NA | NA | [130] |
| Depolymerization Strategy | Depolymerization Products Used as a Feedstock for Fermentation Step | Fermentation Strategy | Products from Fermentation | Titer | Productivity | Yield | Ref. |
|---|---|---|---|---|---|---|---|
| - | Styrene (mock substrate to study upcycling of PS-derived monomer) | Fermentation in shake flask containing 1.85 g/L styrene as a sole carbon source and 67 gN/L NaNH4HPO4·4H2O as a nitrogen source at 30 °C for 48 h by P. putida CA-3 | PHA | NA | NA | 0.099 gPHA/gStyrene | [142] |
| Pyrolysis at 520 °C | Styrene oil (82.8% (w/w) styrene as well as low level of α-methylstyrene, toluene, styrene dimer, and traces of other aromatic compounds) | Fermentation in shake flask containing styrene oil as a sole carbon source and 1 g/L NaNH4HPO4·4H2O as a nitrogen source at 30 °C by P. putida CA-3 | PHA | 0.14 g/L | NA | 0.0625 gPHA/gStyrene oil (0.25 gPHA/gCDW) | [140] |
| Pyrolysis at 520 °C | Styrene oil (82.8% (w/w) styrene as well as low level of α-methylstyrene, toluene, styrene dimer, and traces of other aromatic compounds) | Fermentation in 7.5 L stirred tank reactor feeding a sole carbon source through the gaseous phase contained styrene oil at a concentration of 9.5 mg/L (flow rate 0.15 L/min for the first 3 h of growth and increased to 0.25 L/min for the subsequent 3 h, and finally, to 0.65 L/min for the remainder) at 30 °C by P. putida CA-3 | PHA | 0.32 g/L | NA | 0.1 gPHA/gStyrene oil (0.57 gPHA/gCDW) | [140] |
| Pyrolysis | Distilled styrene oil (89.9% styrene, 5.63% α-methylbenzene, 2.63% toluene, 1.05% ehtylbenzene, 0.43% benzene, 0.19% 1-ethyl-2-methy benzene, and 0.17% unknown) | Fermentation in stirred tank reactor feeding distilled styrene oil at a feed rate of 75 mg/L/h (equivalent to 69 mgC/L/h) and NaNH4HPO4·4H2O at a feed rate of 1.5 mg/L/h at 30 °C by P. putida CA-3 | PHA | 0.82 g/L | NA | 0.28 gPHA/gStyrene oil (0.42 gPHA/gCDW) | [141] |
| - | Styrene (mock substrate to study upcycling of PS-derived monomer) | Fed-batch fermentation in stirred tank reactor feeding styrene as a carbon source overtime through air sparger and NH4Cl as a nitrogen source at different feed rate during the operation period. The fermentation was conducted at 30 °C and pH 6.9 by P. putida CA-3 | mclPHA | 3.36 g/L | NA | 0.32 gPHA/gCDW | [143] |
| Pro-degradation | Pro-degraded PS | Fermentation in shake flask containing 3.7 g/L prodegraded PS emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by C. necator H16 | PHA | 0.52 g/L | NA | 0.39 gPHA/gCDW | [15] |
| Pro-degraded PP was subjected to thermal oxidation (60 °C) in a two-phase system (gas-solid phase) using oxygen-ozone mixture | Thermal oxidized PS (60 °C) | Fermentation in shake flask containing 3.7 g/L thermal oxidized PS emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by C. necator H17 | PHA | 1.72 g/L | NA | 0.48 gPHA/gCDW | [15] |
| Pro-degraded PP was subjected to thermal oxidation (80 °C) in a two-phase system (gas-solid phase) using oxygen-ozone mixture | Thermal oxidized PS (80 °C) | Fermentation in shake flask containing 3.7 g/L thermal oxidized PS emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by C. necator H18 | PHA | 1.28 g/L | NA | 0.42 gPHA/gCDW | [15] |
| Pro-degraded PP was subjected to thermal oxidation (100 °C) in a two-phase system (gas-solid phase) using oxygen-ozone mixture | Thermal oxidized PS (100 °C) | Fermentation in shake flask containing 3.7 g/L thermal oxidized PS emulsified in TSB by sonication as a sole carbon source at 30 °C for 48 h by C. necator H19 | PHA | 0.96 g/L | NA | 0.36 gPHA/gCDW | [15] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lomwongsopon, P.; Varrone, C. Critical Review on the Progress of Plastic Bioupcycling Technology as a Potential Solution for Sustainable Plastic Waste Management. Polymers 2022, 14, 4996. https://doi.org/10.3390/polym14224996
Lomwongsopon P, Varrone C. Critical Review on the Progress of Plastic Bioupcycling Technology as a Potential Solution for Sustainable Plastic Waste Management. Polymers. 2022; 14(22):4996. https://doi.org/10.3390/polym14224996
Chicago/Turabian StyleLomwongsopon, Passanun, and Cristiano Varrone. 2022. "Critical Review on the Progress of Plastic Bioupcycling Technology as a Potential Solution for Sustainable Plastic Waste Management" Polymers 14, no. 22: 4996. https://doi.org/10.3390/polym14224996
APA StyleLomwongsopon, P., & Varrone, C. (2022). Critical Review on the Progress of Plastic Bioupcycling Technology as a Potential Solution for Sustainable Plastic Waste Management. Polymers, 14(22), 4996. https://doi.org/10.3390/polym14224996