Characterization and Comparison of Bioactive Polysaccharides from Grifola frondosa by HPSEC-MALLS-RID and Saccharide Mapping Based on HPAEC-PAD
Abstract
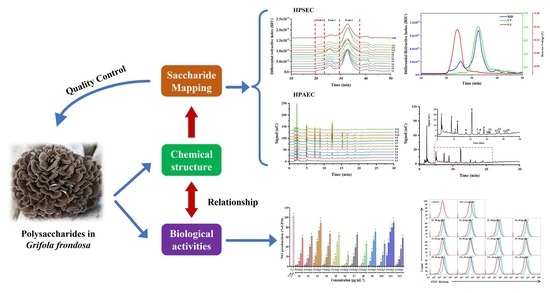
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation of Polysaccharides from G. frondosa
2.3. Monosaccharide Composition of GFPs
2.4. Partial Acid Hydrolysis of GFPs
2.5. Enzymatic Hydrolysis of GFPs
2.6. HPSEC-MALLS-RID Analysis
2.7. HPAEC-PAD Analysis
2.8. Effects of GFPs on Macrophage Functions
2.8.1. Cell Culture
2.8.2. Cell Viability Assay
2.8.3. Nitric Oxide Assay
2.8.4. Phagocytic Activity Test
2.8.5. Determination of Endotoxin Contamination
2.9. Data Analysis and Multivariate Analysis
3. Results and Discussion
3.1. Molecular Weight, Monosaccharides Composition and Content of Polysaccharides from G. frondosa
3.2. Saccharide Mapping based HPSEC-DAD-RID
3.3. Saccharide Mapping Based on HPAEC-PAD
3.3.1. Partial Acid Hydrolysis Fingerprints of GFPs
3.3.2. Enzymatic Hydrolysis Fingerprints of GFPs
3.3.3. HCA of the Saccharide Mapping Based on HPAEC-PAD Chromatogram and Common Peaks
3.4. Effects of Polysaccharides from G. frondosa (GFPs) on Macrophage Functions
3.4.1. Effects of GFPs on Cell Proliferation
3.4.2. Effects of GFPs on Cell Proliferation
3.4.3. Effects of GFPs on Phagocytosis of Macrophages
3.5. HPSEC Fingerprint-Immune Activity Relationship
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Liu, Y.; Sarker, M.M.R.; Yan, X.; Yang, C.; Zhao, L.; Lv, X.; Liu, B.; Zhao, C. Structural characterization and antidiabetic potential of a novel heteropolysaccharide from Grifola frondosa via IRS1/PI3K-JNK signaling pathways. Carbohydr. Polym. 2018, 198, 452–461. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, X.; Hao, C.; Zeng, P.; Zhang, M.; Liu, Y.; Chang, Y.; Zhang, L. Grifola frondosa polysaccharide: A review of antitumor and other biological activity studies in China. Discov. Med. 2018, 25, 159–176. [Google Scholar] [PubMed]
- Guan, J.; Li, S.P. Discrimination of polysaccharides from traditional Chinese medicines using saccharide mapping-Enzymatic digestion followed by chromatographic analysis. J. Pharm. Biomed. Anal. 2010, 51, 590–598. [Google Scholar] [CrossRef]
- Li, S.P.; Wu, D.T.; Lv, G.P.; Zhao, J. Carbohydrates analysis in herbal glycomics. Trac-Trends Anal. Chem. 2013, 52, 155–169. [Google Scholar] [CrossRef]
- Deng, Y.; Chen, L.X.; Zhu, B.J.; Zhao, J.; Li, S.P. A quantitative method for polysaccharides based on endo-enzymatic released specific oligosaccharides: A case of Lentinus edodes. Int. J. Biol. Macromol. 2022, 205, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Zhao, J.; Feng, K.; Hu, D.J.; Li, S.P. Comparison and characterization of polysaccharides from natural and cultured Cordyceps using saccharide mapping. Anal. Bioanal. Chem. 2011, 399, 3465–3474. [Google Scholar] [CrossRef]
- Wu, D.T.; Cheong, K.L.; Wang, L.Y.; Lv, G.P.; Ju, Y.J.; Feng, K.; Zhao, J.; Li, S.P. Characterization and discrimination of polysaccharides from different species of Cordyceps using saccharide mapping based on PACE and HPTLC. Carbohydr. Polym. 2014, 103, 100–109. [Google Scholar] [CrossRef]
- Cheong, K.L.; Wu, D.T.; Hu, D.J.; Zhao, J.; Cao, K.Y.; Qiao, C.F.; Han, B.X.; Li, S.P. Comparison and Characterization of the Glycome of Panax Species by High-Performance Thin-Layer Chromatography. JPC—J. Planar Chromatogr. -Mod. TLC 2014, 27, 449–453. [Google Scholar] [CrossRef]
- Zhu, B.J.; Yan, Z.Y.; Hong, L.; Li, S.P.; Zhao, J. Quality evaluation of Salvia miltiorrhiza from different geographical origins in China based on qualitative and quantitative saccharide mapping and chemometrics. J. Pharm. Biomed. Anal. 2020, 191, 113583. [Google Scholar] [CrossRef]
- Mechelke, M.; Herlet, J.; Benz, J.P.; Schwarz, W.H.; Zverlov, V.V.; Liebl, W.; Kornberger, P. HPAEC-PAD for oligosaccharide analysis—Novel insights into analyte sensitivity and response stability. Anal. Bioanal. Chem. 2017, 409, 7169–7181. [Google Scholar] [CrossRef]
- Otieno, D.O.; Ahring, B.K. A thermochemical pretreatment process to produce xylooligosaccharides (XOS), arabinooligosaccharides (AOS) and mannooligosaccharides (MOS) from lignocellulosic biomasses. Bioresour. Technol. 2012, 112, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.S.; Jane, J. Effects of Pushing Agents on the Separation and Detection of Debranched Amylopectin by High-Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection. J. Liq. Chromatogr. 1995, 18, 63–80. [Google Scholar] [CrossRef]
- White, D.R.; Hudson, P.; Adamson, J.T. Dextrin characterization by high-performance anion-exchange chromatography-pulsed amperometric detection and size-exclusion chromatography-multi-angle light scattering-refractive index detection. J. Chromatogr. A 2003, 997, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Pöhnl, T.; Böttcher, C.; Schulz, H.; Stürtz, M.; Widder, S.; Carle, R.; Schweiggert, R.M. Comparison of high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) and ultra-high performance liquid chromatography with evaporative light scattering (UHPLC-ELSD) for the analyses of fructooligosaccharides in onion (Allium cepa L.). J. Food Compos. Anal. 2017, 63, 148–156. [Google Scholar] [CrossRef]
- Rohrer, J.S.; Townsend, R.R. Separation of partially desialylated branched oligosaccharide isomers containing α (2←3)- and α (2←6)-linked Neu5Ac. Glycobiology 1995, 5, 391–395. [Google Scholar] [CrossRef]
- Yu, J.; Ji, H.Y.; Liu, C.; Liu, A.J. The structural characteristics of an acid-soluble polysaccharide from Grifola frondosa and its antitumor effects on H22-bearing mice. Int. J. Biol. Macromol. 2020, 158, 1288–1298. [Google Scholar] [CrossRef]
- Wu, D.T.; Xie, J.; Wang, L.Y.; Ju, Y.J.; Lv, G.P.; Leong, F.; Zhao, J.; Li, S.P. Characterization of bioactive polysaccharides from Cordyceps militaris produced in China using saccharide mapping. J. Funct. Food. 2014, 9, 315–323. [Google Scholar] [CrossRef]
- Wu, J.Y.; Siu, K.C.; Geng, P. Bioactive Ingredients and Medicinal Values of Grifola frondosa (Maitake). Foods 2021, 10, 95. [Google Scholar] [CrossRef]
- Cheong, K.L.; Wu, D.T.; Zhao, J.; Li, S.P. A rapid and accurate method for the quantitative estimation of natural polysaccharides and their fractions using high performance size exclusion chromatography coupled with multi-angle laser light scattering and refractive index detector. J. Chromatogr. A 2015, 1400, 98–106. [Google Scholar] [CrossRef]
- Wu, D.T.; Lam, S.C.; Cheong, K.L.; Wei, F.; Lin, P.C.; Long, Z.R.; Lv, X.J.; Zhao, J.; Ma, S.C.; Li, S.P. Simultaneous determination of molecular weights and contents of water-soluble polysaccharides and their fractions from Lycium barbarum collected in China. J. Pharm. Biomed. Anal. 2016, 129, 210–218. [Google Scholar] [CrossRef]
- Zimm, B.H. Apparatus and methods for measurement and interpretation of the angular variation of light scattering; preliminary results on polystyrene solutions. J. Chem. Phys. 1948, 16, 1099–1116. [Google Scholar] [CrossRef]
- Miranda, K.M.; Espey, M.G.; Wink, D.A. A Rapid, Simple Spectrophotometric Method for Simultaneous Detection of Nitrate and Nitrite. Nitric Oxide 2001, 5, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Wu, D.T.; Xie, J.; Zhao, J.; Li, S.P. Characterization and comparison of bioactive polysaccharides from the tubers of Gymnadenia conopsea. Food Hydrocoll. 2015, 43, 199–206. [Google Scholar] [CrossRef]
- He, X.R.; Wang, X.X.; Fang, J.C.; Chang, Y.; Ning, N.; Guo, H.; Huang, L.H.; Huang, X.Q.; Zhao, Z.F. Polysaccharides in Grifola frondosa mushroom and their health promoting properties: A review. Int. J. Biol. Macromol. 2017, 101, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Wei, S.X.; Huang, H. Recent advances in Grifola frondosa polysaccharides: Production, properties, and bioactivities. Curr. Opin. Food Sci. 2022, 49, 100946. [Google Scholar] [CrossRef]
- Cui, F.J.; Tao, W.Y.; Xu, Z.H.; Guo, W.J.; Xu, H.Y.; Ao, Z.H.; Jin, J.; Wei, Y.Q. Structural analysis of anti-tumor heteropolysaccharide GFPS1b from the cultured mycelia of Grifola frondosa GF9801. Bioresour. Technol. 2007, 98, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, W.; Zhu, Y.; Chen, Y.; Zhang, W.; Yu, P.; Mao, G.; Zhao, T.; Feng, W.; Yang, L.; et al. Structural elucidation and antioxidant activity a novel Se-polysaccharide from Se-enriched Grifola frondosa. Carbohydr. Polym. 2017, 161, 42–52. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, J.; Ni, X.; Li, J.; Liu, Q.; Dong, Q.; Duan, J.; Ding, K. Inducement of Cytokine Release by GFPBW2, a Novel Polysaccharide from Fruit Bodies of Grifola frondosa, through Dectin-1 in Macrophages. J. Agric. Food Chem. 2013, 61, 11400–11409. [Google Scholar] [CrossRef]
- Hu, D.J.; Cheong, K.L.; Zhao, J.; Li, S.P. Chromatography in characterization of polysaccharides from medicinal plants and fungi. J. Sep. Sci. 2013, 36, 1–19. [Google Scholar] [CrossRef]
- Aktan, F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004, 75, 639–653. [Google Scholar] [CrossRef]






| Code | Regions | Peak 1 | Peak 2 | Peak 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mw×106 /Da | Mw/Mn | Content/% | Mw×106 /Da | Mw/Mn | Content/% | Mw×105/ Da | Mw/Mn | Content/% | ||
| S1 | Zhejiang Province | 5.06 | 1.06 | 1.50 | 3.88 | 1.41 | 14.75 | 1.22 | 1.77 | 62.75 |
| S2 | Zhejiang Province | 5.97 | 1.08 | 1.75 | 3.70 | 1.32 | 13.25 | 1.28 | 1.72 | 59.90 |
| S3 | Zhejiang Province | 6.78 | 1.02 | 1.46 | 3.29 | 1.38 | 12.60 | 1.24 | 1.64 | 53.70 |
| S4 | Zhejiang Province | 7.35 | 1.04 | 0.65 | 3.72 | 1.36 | 11.86 | 1.26 | 1.71 | 44.14 |
| S5 | Zhejiang Province | 7.01 | 1.02 | 1.00 | 3.11 | 1.49 | 9.90 | 1.08 | 1.52 | 42.55 |
| S6 | Zhejiang Province | 6.80 | 1.05 | 1.15 | 3.18 | 1.47 | 7.30 | 1.23 | 1.57 | 36.70 |
| S7 | Zhejiang Province | 6.87 | 1.03 | 1.65 | 2.63 | 1.59 | 7.85 | 1.01 | 1.55 | 49.05 |
| S8 | Hebei Province | 5.51 | 1.09 | 1.19 | 3.69 | 1.59 | 18.86 | 1.11 | 1.70 | 57.74 |
| S9 | Fujian Province | 8.71 | 1.03 | 1.50 | 3.84 | 1.42 | 10.30 | 1.39 | 1.61 | 43.50 |
| S10 | Fujian Province | 5.00 | 1.09 | 1.30 | 3.26 | 1.36 | 9.50 | 1.11 | 1.60 | 43.60 |
| S11 | Hunan Province | 5.48 | 1.02 | 1.90 | 5.64 | 1.38 | 9.15 | 1.51 | 1.66 | 66.00 |
| S12 | Hunan Province | 8.49 | 1.06 | 2.10 | 4.17 | 1.36 | 9.30 | 1.21 | 1.61 | 61.40 |
| Code | HPSEC | HPAEC-PAD | |||
|---|---|---|---|---|---|
| GFPs | Partial acid | α-amylase | Cellulase | β-1,3-glucanase | |
| S1 | 0.990 | 0.954 | 0.400 | 0.843 | 0.912 |
| S2 | 0.994 | 0.992 | 0.754 | 0.968 | 0.957 |
| S3 | 0.990 | 0.996 | 0.835 | 0.939 | 0.985 |
| S4 | 0.981 | 0.995 | 0.651 | 0.980 | 0.999 |
| S5 | 0.999 | 0.981 | 0.934 | 0.955 | 0.998 |
| S6 | 0.996 | 0.954 | 0.924 | 0.925 | 0.992 |
| S7 | 0.995 | 0.989 | 0.981 | 0.991 | 0.781 |
| S8 | 0.990 | 0.996 | 0.969 | 0.970 | 0.998 |
| S9 | 0.997 | 0.989 | 0.922 | 0.994 | 0.954 |
| S10 | 0.992 | 0.984 | 0.912 | 0.980 | 0.978 |
| S11 | 0.991 | 0.846 | 0.391 | 0.699 | 0.713 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.; Zhang, W.; Zhao, J.; Chen, B.; Liu, F.; Li, S. Characterization and Comparison of Bioactive Polysaccharides from Grifola frondosa by HPSEC-MALLS-RID and Saccharide Mapping Based on HPAEC-PAD. Polymers 2023, 15, 208. https://doi.org/10.3390/polym15010208
Zhu B, Zhang W, Zhao J, Chen B, Liu F, Li S. Characterization and Comparison of Bioactive Polysaccharides from Grifola frondosa by HPSEC-MALLS-RID and Saccharide Mapping Based on HPAEC-PAD. Polymers. 2023; 15(1):208. https://doi.org/10.3390/polym15010208
Chicago/Turabian StyleZhu, Baojie, Wenxia Zhang, Jing Zhao, Bilian Chen, Fei Liu, and Shaoping Li. 2023. "Characterization and Comparison of Bioactive Polysaccharides from Grifola frondosa by HPSEC-MALLS-RID and Saccharide Mapping Based on HPAEC-PAD" Polymers 15, no. 1: 208. https://doi.org/10.3390/polym15010208
APA StyleZhu, B., Zhang, W., Zhao, J., Chen, B., Liu, F., & Li, S. (2023). Characterization and Comparison of Bioactive Polysaccharides from Grifola frondosa by HPSEC-MALLS-RID and Saccharide Mapping Based on HPAEC-PAD. Polymers, 15(1), 208. https://doi.org/10.3390/polym15010208